Part:BBa_K1060000
AroG; DAHP synthase
The aroG gene, coding for 3-deoxy-D-arabino-heptulosonate-7-phosphate synthase, is a part of the Shikimate pathway. It converts phosphoenolpyruvate and erythrose-4P to chorismate. In our project chorismate was one of the precursors needed for the biosynthesis of methyl salicylate (see also BBa_K1060003). As basic chorismate levels inside E. coli are limited. We wanted to upregulate the biosynthesis of chorismate. The basic aroG gene can be repressed by phenylalanine. However, mutatations in the amino acid sequence (Pro150Leu and Leu175Asp) can prevent this repression by phenylalanine.
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21INCOMPATIBLE WITH RFC[21]Illegal XhoI site found at 934
- 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000INCOMPATIBLE WITH RFC[1000]Illegal SapI.rc site found at 241
2021 XJTU-China's Improvement
This part has been improved by 2021 team: XJTU-China. The improved version, AroG-S211F(BBa_K3832000, has been analyzed by structure modelling and measured.
Here some important results of our improvement have been shown below. And for more detailed methods and results, please check pages of Part:BBa_K3832000 and our wiki Team:XJTU-China/Improve
Structural Modelling Result
According to the docking results, the protein pocket conformation of wild-type and mutant AroG bound to PEP has not changed significantly. The Red is the experimentally measured conformation of Phe in the crystal protein bound by aroG and Phe, and the pink is the docking site and the conformation of Phe predicted by AutoDockTools software. In general, the changes in binding PEP sites are not very significant.
Phe site:
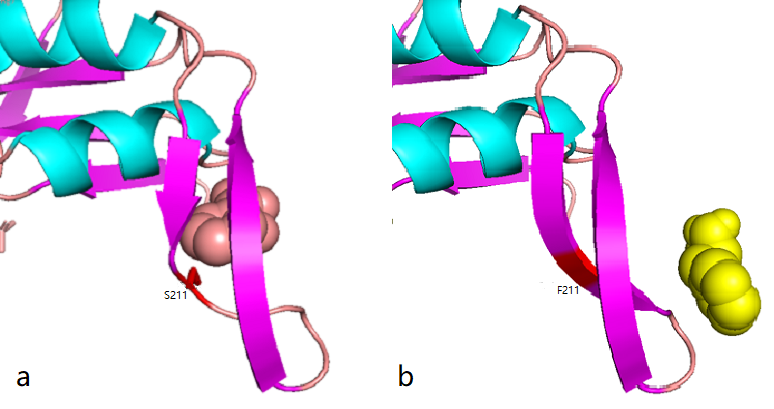
Figure 2.8 Comparison between docking site of Phe, (a)Binding site of wild-type AroG and Phe;(b)Binding site of mutant AroG-S211F and Phe, in which mutant site is shown as red
It can be seen from the docking results that the Ser at AroG 211 mutating to Phe changes the conformation of the protein pocket which originally binds to Phe, and the binding site of Phe changes accordingly. The binding energy calculated by AutoDockTools software shows that the binding ability of mutant aroG and Phe becomes weaker. Therefore, it can be concluded that the conformation of the mutant aroG and Phe binding protein pocket changes, so that the binding ability of Phe to it becomes smaller, and the allosteric inhibition effect of Phe is reduced, finally the catalytic efficiency of aroG on PEP increases.
Characterization & Measurement
| None |
