Part:BBa_K3887007
Fre-L3-PyrH
Fre-L3-PyrH is an fusion enzyme of Fre and PyrH using linker L3.
Description
PyrH is another type of halogenase enzyme which adds halogen element bromine or chlorine at C5 during conversion of indole[1]. In our project, it is used to biosynthesis other types of indigo, similar to SttH producing Tyrian Purple. It is also applied as fusion enzyme with Fre-L3 for better solubility and then cloned to pACYC backbone as Figure 1[1,2].
Experiments and Results
We constructed halogenase plasmids using Gibson Assembly and verified by colony PCR and gene sequencing . According to the results in Figure 2, we can roughly identify Fre-L3-PyrH gene band obviously comparing with DNA Marker. And then the PCR product fragment was sent for sequencing and the results were also consistent with expected.
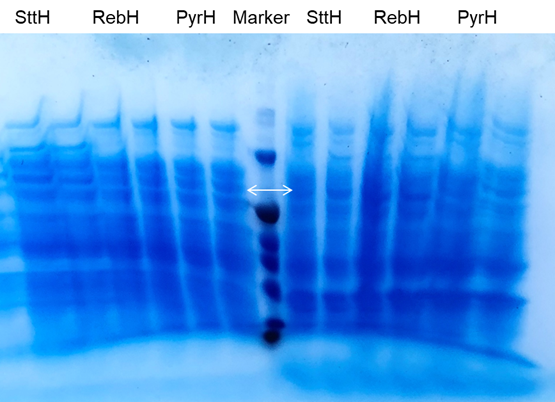
Next, we started the protein level experiment to test gene expression. For better expression of enzymes, we optimize the induction condition of gene expression: three different temperatures (18℃, 30℃, 37℃) and inducer concentrations (0 mM, 0.1mM, 1mM) were set for test.the collected cells after induction were disrupted, both of the pellet and supernatant were used for SDS-PAGE detection (Figure 3). From the results of three SDS-PAGE electrophoresis gels, it can be seen that compared with the control group induced by 0mM IPTG, there are obvious changes in the lanes induced by IPTG, which proves Fre-L3-PyrH has been effectively expressed under the induction. Through the expression of halogenase at different temperatures under the same IPTG concentration, we can get the following conclusions: PyrH protein expressed better and has obvious bands at 18℃ compared to the expression at 30℃ and 37℃, so we can conclude that 18°C is the most suitable induction temperature for PyrH.
However, it is not difficult to see that the content of Fre-L3-PyrH expressed in the supernatant solution is low. This may be due to low solublity of halogenases, even through Fre-L3 protein connection, the solubility is still not effectively improved, most enzymes are still exsit in the precipitation. It may also be related to our parameter settings in the cell disruption. So we subsequently performed the protein expression induction experiment again with optimized induction conditions and ultrasonication strength, and better results were obtained (Figure 4).
Finally, we did a fermentation experiment with E.coli BL21 containing Fre-L3-PyrH and the results were shown in Figure 5, from which we can see different indigo derivatives as expected.
References
[1]Lee, J., Kim, J., Song, J.E. et al. Production of Tyrian purple indigoid dye from tryptophan in Escherichia coli. Nat Chem Biol 17, 104–112 (2021)
[2]Alfieri A, Malito E, Orru R, Fraaije MW, Mattevi A. Revealing the moonlighting role of NADP in the structure of a flavin-containing monooxygenase. Proc Natl Acad Sci U S A. 2008 May 6;105(18):6572-7 Sequence and Features
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12INCOMPATIBLE WITH RFC[12]Illegal NotI site found at 2136
- 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25INCOMPATIBLE WITH RFC[25]Illegal NgoMIV site found at 943
Illegal NgoMIV site found at 1561 - 1000INCOMPATIBLE WITH RFC[1000]Illegal BsaI site found at 2116
Illegal BsaI.rc site found at 2086
Illegal SapI site found at 1158
Illegal SapI.rc site found at 301
Illegal SapI.rc site found at 421
| None |