Part:BBa_K3798015
Status: 500
Content-type: text/html
Software error:
Can't call method "errstr" on an undefined value at /websites/parts.igem.org/cgi/lib/BBDB.pm line 35. Compilation failed in require at /websites/parts.igem.org/cgi/lib/Plate.pm line 13. BEGIN failed--compilation aborted at /websites/parts.igem.org/cgi/lib/Plate.pm line 13. Compilation failed in require at /websites/parts.igem.org/cgi/lib/Well.pm line 11. BEGIN failed--compilation aborted at /websites/parts.igem.org/cgi/lib/Well.pm line 11. Compilation failed in require at /websites/parts.igem.org/cgi/lib/Blast.pm line 15. BEGIN failed--compilation aborted at /websites/parts.igem.org/cgi/lib/Blast.pm line 15. Compilation failed in require at /websites/parts.igem.org/cgi/lib/Sample.pm line 24. BEGIN failed--compilation aborted at /websites/parts.igem.org/cgi/lib/Sample.pm line 24. Compilation failed in require at /websites/parts.igem.org/cgi/lib/WrapPart.pm line 10. BEGIN failed--compilation aborted at /websites/parts.igem.org/cgi/lib/WrapPart.pm line 10. Compilation failed in require at /websites/parts.igem.org/cgi/lib/BBWeb.pm line 12. BEGIN failed--compilation aborted at /websites/parts.igem.org/cgi/lib/BBWeb.pm line 12. Compilation failed in require at /websites/parts.igem.org/cgi/lib/Change.pm line 8. BEGIN failed--compilation aborted at /websites/parts.igem.org/cgi/lib/Change.pm line 8. Compilation failed in require at /websites/parts.igem.org/cgi/lib/Part.pm line 12. BEGIN failed--compilation aborted at /websites/parts.igem.org/cgi/lib/Part.pm line 12. Compilation failed in require at /websites/parts.igem.org/cgi/partsdb/putout.cgi line 8. BEGIN failed--compilation aborted at /websites/parts.igem.org/cgi/partsdb/putout.cgi line 8.
For help, please send mail to this site's webmaster, giving this error message and the time and date of the error.
The CBD domains of the protein is derived from the previous part of BBa_K1934020. The eGFP domain of the protein can be the signal of cellulose attachment and can be a subject of modeling. We use this protein to demonstrate the ability of using CBD domains to bind to the cellulose.
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000COMPATIBLE WITH RFC[1000]
Charcterization
Connection between CBD-eGFP and cellulose
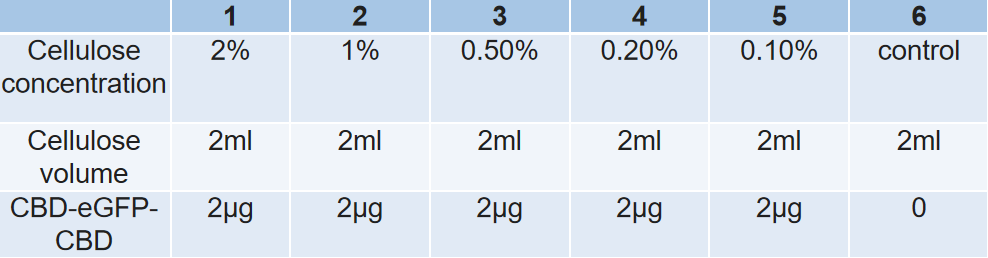
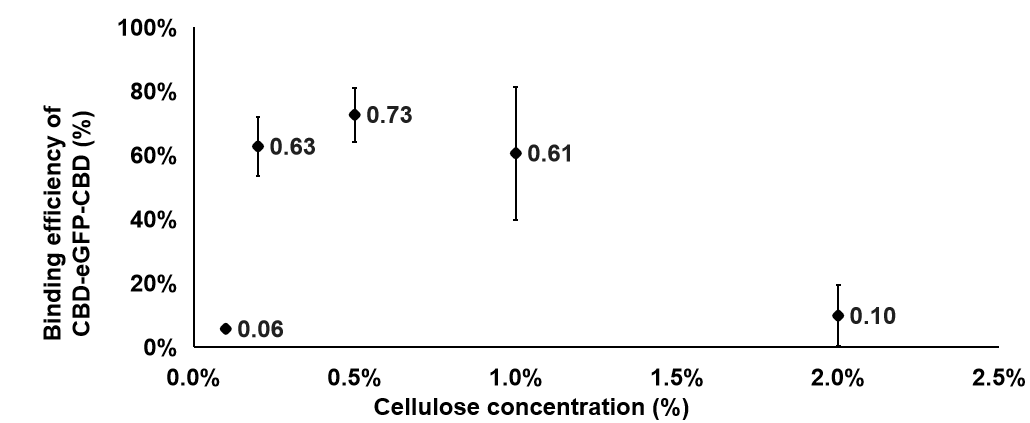
1. In this experiments, we hoped to find out the best condition for the binding between cellulose and CBD-SA. Firstly, we intended to find out how the concentration of cellulose affects the binding efficiency between cellulose and CBD-SA. According to the given cultivation environment above, 6 groups of different cellulose concentrations were set up, and the fluorescent value of each group was tested in the aim of calculating binding efficiency between Cellulose and CBD-SA.
Samples with different concentration of cellulose.
CBD-eGFP was used to replace CBD-SA in this experiment, because it has similar properties with CBD-SA and was designed as a visible fluorescent marker. Precipitate will form if cellulose binds the protein. That provided us a better way to exam the concentration of protein by detecting the fluorescent value. The fluorescent value of the supernatant in the solution was detected. The way we calculate the binding efficiency was as following: Firstly, we used the fluorescent value of supernatant in solution minus the fluorescence that already in the solution to get the remaining fluorescent value after the binding. Then, we used the total value minus the value we got and it refers to the binding value. The binding efficiency is the value which binding value divided by total fluorescence.
The formula is Binding efficiency = [Total fluorescence - (Remaining fluorescence - Background value)]/Total value
We can learn from the graph that the binding efficiency is relatively high when the concentration of cellulose is at 0.2% and 0.5%. Considering the cellulose will be sticky when the concentration is too high, we took 0.2% as our finally concentration.
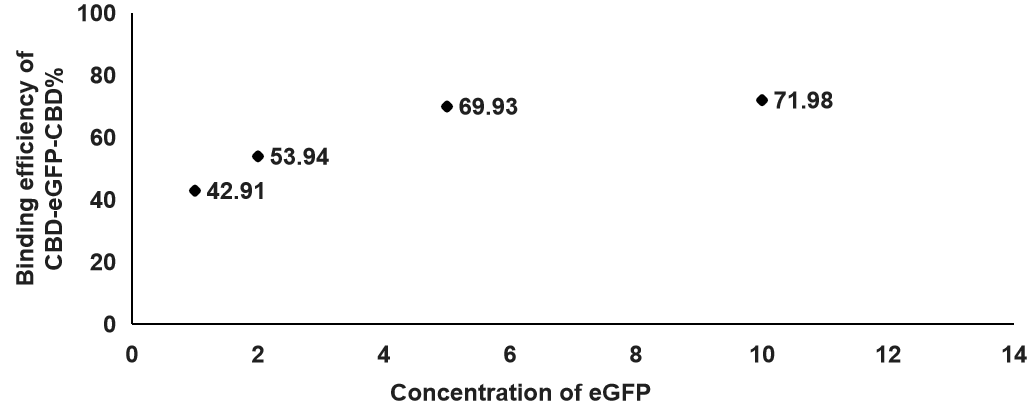
2. After we determined which concentration of cellulose is best for binding, we also tested the optimal eGFP concentration for the binding efficiency between CBD-SA and cellulose. Additionally, the concentration of cellulose in this experiment is at 0.2%.
According to this figure, we found out that the binding efficiency was the highest when the concentration of eGFP was at 10, which means the binding efficiency was increasing when the concentration of eGFP was higher. That will be the concentration we use in the following experiments.
| None |