Part:BBa_K4035002
Dimerization of the copper metallothionein 1 : CUP1-(GGGGS)3-CUP1
This protein is made of two copies of the yeast copper metallothionein protein, CUP1 (BBa_M45090), linked together by a flexible linker made of three times the GGGGS amino acids sequence.
Usage and Biology
Copper metallothionein CUP1 (BBa_M45090) is a protein responsible for copper binding in the yeast Saccharomyces cerevisiae. In order to increase the copper retrieval efficiency, two copies of CUP1 have been linked together by the (GGGGS)3 aa sequence. GGGGS is a common flexible linker and was inserted 3 times in order to allow distances between the two copies of CUP1 and avoid interaction. This fusion protein was expressed on the outer membrane of S. cerevisiae by insertion of this part sequence into the pCTcon2V5 (1) plasmid. The expression system (BBa_K4035009) consisted of fusing the dimer on a yeast surface display (1) under the control of the Gal1 promoter. This resulted of the production of a fusion protein Aga2-CUP1-(GGGGS)3-CUP1-V5.
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21INCOMPATIBLE WITH RFC[21]Illegal BamHI site found at 190
- 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000COMPATIBLE WITH RFC[1000]
Characterization
Expression of the protein in S.cerevisiae
Western Blot Analysis
In order to characterize the protein expression, two experiments were performed, the first being a Western Blot analysis. After having transformed the EBY100 yeast with our newly formed plasmid pCTcon2V5-CUP1-(GGGGS)3-CUP1-V5, we analyzed its protein expression. For control we also tested the wild type yeast (untransformed) as well as a transformed yeast with the plasmid backbone (without insert) in SGCAA, a medium containing galactose allowing for protein expression through the activation of the Gal1 promoter. As the plasmid contains a Gal1 promoter, the system can only be expressed in the presence of galactose.
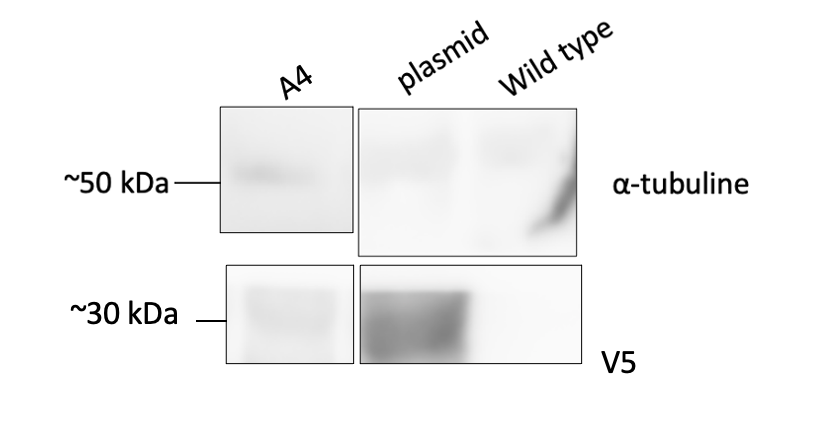
Figure 1 : The first lane (A4) is the yeast transformed with our plasmid containing the CUP1 dimer and the second and third lanes are, respectively, EBY100 and pCTcon2V5. EBY100 is the wild type yeast and serves as a negative control whereas pCTcon2V5 is the transformed yeast with only the plasmid backbone and serves as a positive control.
Wild type yeast shows no presence of the V5 tag, as expected, while plasmid bakcbone has signal which proves that our system is expressed in the transformed yeast when induced with galactose. Our transformed yeast with the Aga2-CUP1-(GGGGS)3-CUP1-V5 protein also shows expression of the system.
We can see that we have two bands on the gel, one is approximately 30 kDa which is the size of the fusion protein Aga2-CUP1-(GGGGS)3-CUP1-V5 and the other is slightly smaller. The smaller band could be a truncated version of the protein since we identified a second in-frame start codon in the DNA sequence.
The presence of our CUP1 dimer in our yeast transformants is thus shown.
Immunocytochemistry Assay
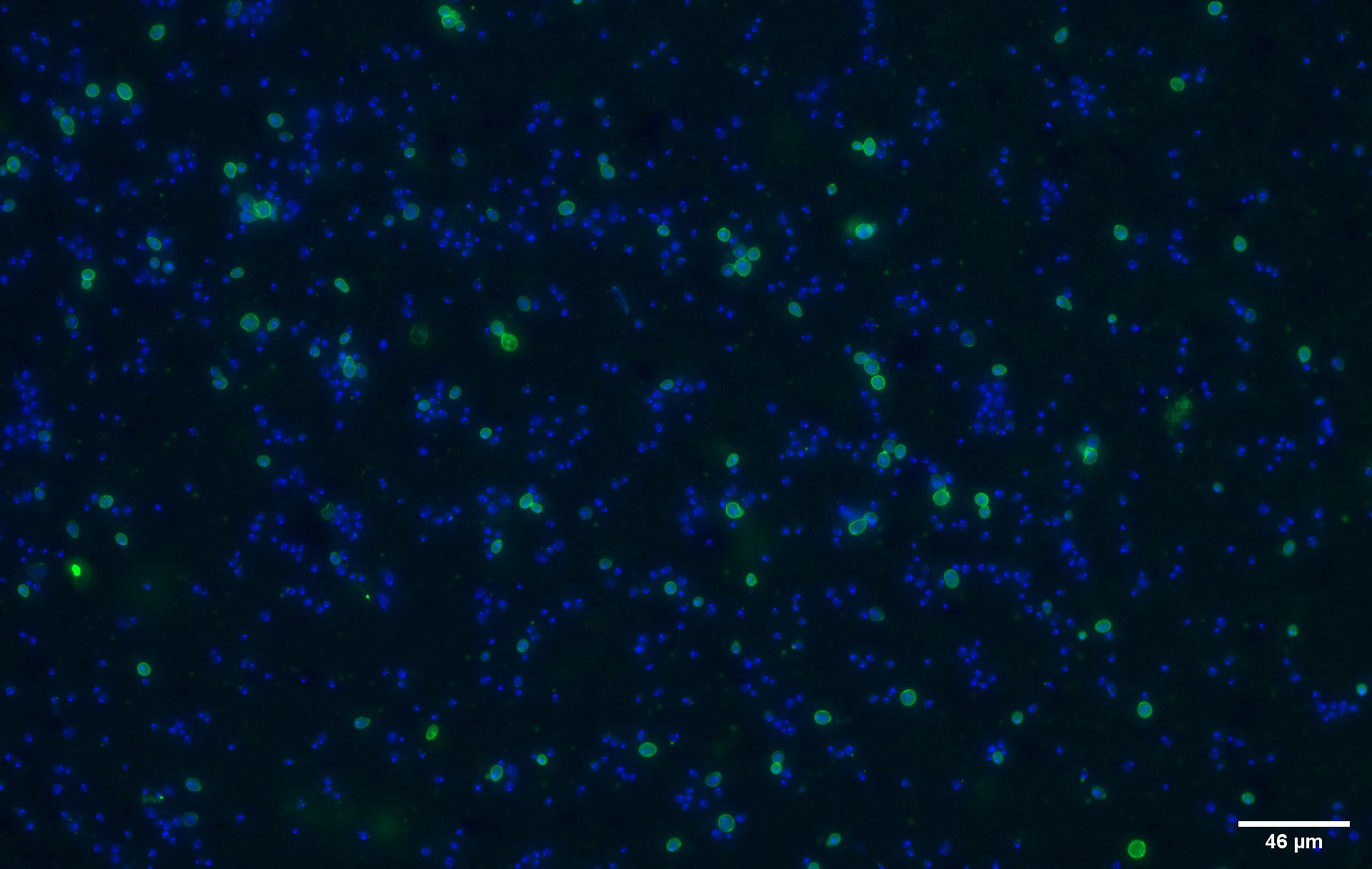
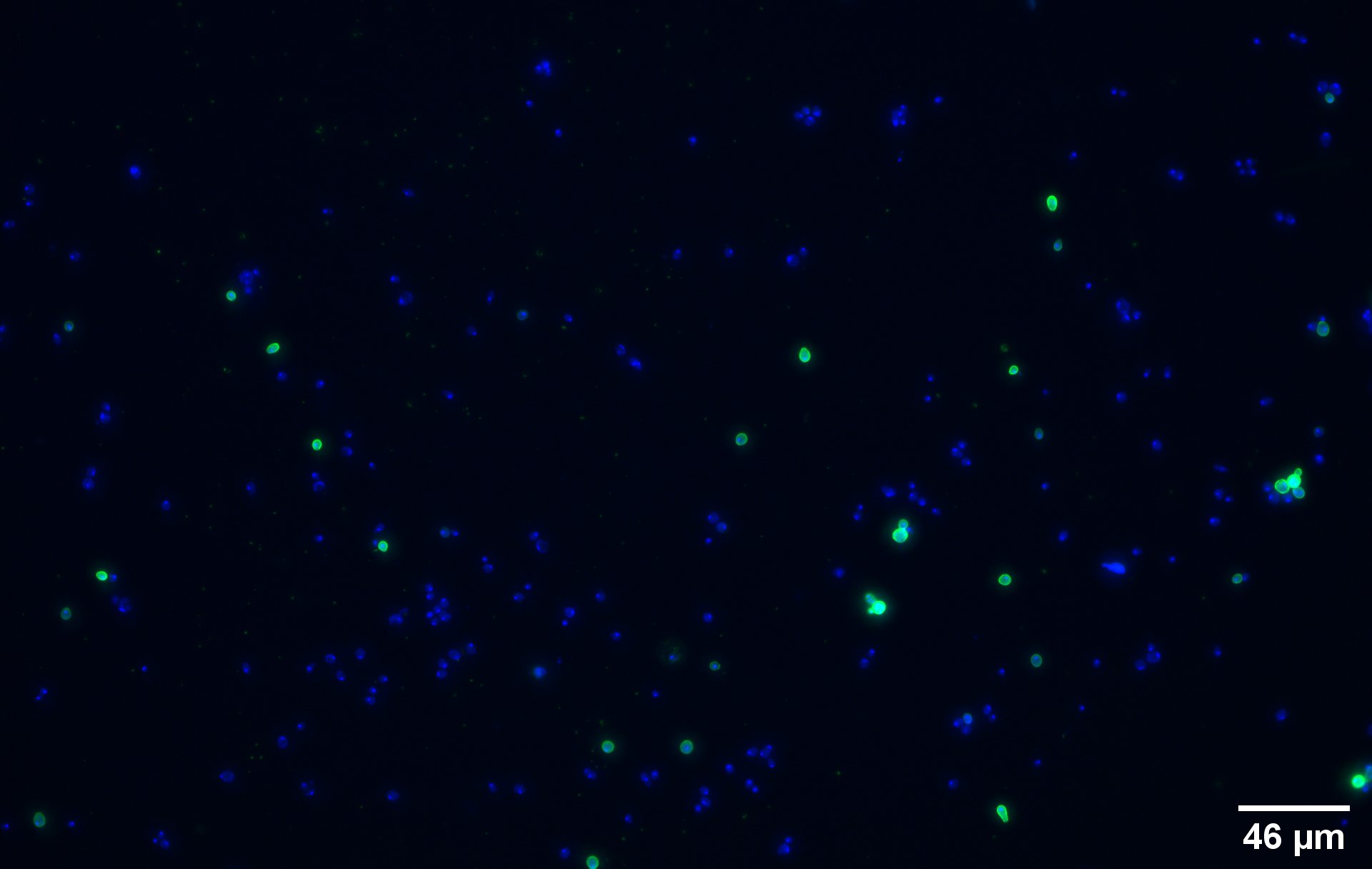
To show that the fusion protein is expressed at the membrane of the cell we performed an Immunocytochemistry assay. The cells were incubated with a primary mouse anti-V5 antibody as well as with a secondary goat anti-mouse couple with Alexa Fluor 488. The same strains for control have been used namely, plasmid backbone induced (Figure 2c) and wild type yeast (Figure 2d).
The nuclei have been stainined with DAPI and are depicted in blue on the figures. The green disks are representing the the recombinant yeast cells expressing the CUP1 dimer at their surface (Figure 2a).
Due to the more intense circle we can clearly see that our system is expressed on the membrane of the protein (better seen on Figure 2b which is a higher magnification picture of yeast transformed with a single copy of CUP1, part BBa_K4035001).
We can also remark that not all the cells are expressing the system. This is because the expression is not 100% efficient, concordant to published expression system (1)..
On the negative control, the little green signal we see is background noise or unspecific antibody binding.

Growth and Survival Characterization
Growth curves of the different yeast strains
In order to check if our expression system would affect the growth of our micro-organism, we performed multiple growth curves with different parameters.
Colony forming assays of the different yeast strains
A second experiment was done to check if the expression system would affect the growth of the yeast cells, a colony forming assay. That experiment consisted of counting the colonies formed by the different yeast strains, respectively wild type, backbone uninduced, backbone induced and transformant, on agar plates.
Copper absorption assays
To finally test our system we imagined an experiment to measure how much copper our transformed yeast could absorb.
//cds/membrane/extracellular
//chassis/eukaryote/yeast
| biology | CUP1 |
| chassis | S.cerevisiae |
| control | pCTcon2V5, WT EBY100 |
| direction | Forward |
| function | copper binding protein |