Part:BBa_K3717009
α-Galactosidase with T7 + RBS, N-Terminal 6x His-Tag, and Double Terminator
The composite part utilizes a T7 promoter + RBS (BBa_K525998), α-Galactosidase (BBa_K3717006), and double terminator (BBa_B0015).

Figure 1. α-Galactosidase with Φ29 DNA polymerase with T7 promoter, RBS and double terminator construct
Construct Design
We derived the sequence of α-Galactosidase from Bacteroides fragilis and optimized the sequence for E. coli protein expression. We then attached a 6x Histidine Tag (6x His-Tag) upstream of the α-Galactosidase sequence followed by a glycine-serine linker (GS linker) to form our open reading frame (ORF) (BBa_K3717006) for purification purposes. We flanked our open reading frame with a T7 promoter + RBS (BBa_K525998) upstream of the open reading frame and a double terminator (BBa_B0015) downstream of the sequence. This composite part (BBa_K3717009) was assembled through DNA synthesis by IDT.
Characterization
Protein Expression and Purification
We tested protein expression of the composite parts by transforming our plasmids into BL21(DE3) E. coli cells. We grew an overnight culture of the BL21 cells with our plasmids then diluted our cells to a standardized OD600 of ~0.1 and let it grow until an OD600 of 0.5~0.6. The diluted cultures of OD600 0.5~0.6 were then induced for expression with 0.5 M IPTG stock (to a final concentration of 0.5mM in the culture) and allowed to grow and induce overnight at room temperature.
We harvested the cells after the overnight induction and lysed them either through sonication or with xTractor Lysis Buffer spiked with 500mM Imidazole stock (to a final concentration of 20mM in the lysate solution) [2]. We purified the Histidine tagged proteins using Ni sepharose affinity chromatography [2]. We then utilized SDS-PAGE to confirm the sizes of purified proteins.
Our results indicate a protein band at roughly 69.7 kDa, which is the molecular weight of our α-Galactosidase enzyme with the 6x His tag and GS linker attached, proving that our α-Galactosidase (Part: BBa_K3717009) was expressed and purified.
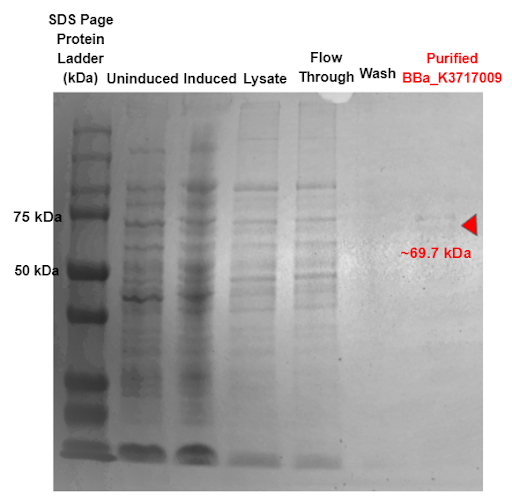
Figure 2. SDS-PAGE of purified proteins with the T7 promoter α-Galactosidase expressing construct (BBa_K3717009). Red triangles indicate expected size for the part.
Functional Testing
Colorimetric Tests
We buffer exchanged α-Galactosidase (α-Gal) into 0.1 M Sodium Phosphate buffer (pH 7.4) by dialysis and concentrated the solution to 0.5 mL using centrifugal ultrafiltration. We confirmed the functionality of the enzyme using 3 assays: colorimetric tests, thin-layer chromatography, and mass spectroscopy.
To test the efficiency and efficacy of the enzyme, we injected the enzyme into a solution containing a colorimetric substrate that imitated the structure of an A antigen, with the product creating a color that can be measured using a 405 nm absorbance.
A colorimetric substrate for α-Galactosidase (α-Gal) was obtained from Sigma Aldrich (Fig 3). This substrate contains a 4-nitrophenol leaving group, which turns yellow upon successful cleavage in solution. The concentration of 4-nitrophenol was quantified at absorbance 405 nm using a 96 well plate based assay [3].

Figure 3. Colorimetric substrate for α-Gal.
We performed small-scale colorimetric tests to verify the function of the enzyme. To each well, we added 50 μL of 10 mM substrate, 10 μL of enzyme, 30 μL of water, and 10 μL of 10x Glycobuffer 1 (50 mM CaCl2, 500 mM sodium acetate, pH 5.5), a buffer recommended by New England Biolabs to ensure optimal enzyme activity [4]. Following 2 hours of reaction at room temperature, we diluted the well contents in 1.9 mL of water and took absorbance readings at 405 nm. Results indicate that α-Gal successfully cleaved its substrates (Fig 4). Moreover, we demonstrated the specificity of the enzyme through the lack of enzyme activity on the other substrate.

Figure 4. Small-scale colorimetrics tests verify the functionality and specificity of our purified enzymes. a) 96-well plates following enzyme reaction. b) absorbance readings of diluted well contents. c) buffers and solutions used in substrate and control wells.
To quantify the activity of the enzyme, we performed enzyme reactions at various substrate concentrations and measured the absorbance at different time intervals over a constant time period (Fig 5). Results from at least 3 independent trials for each enzyme were averaged and used to calculate Michaelis–Menten constants for enzyme efficiency (see enzyme model in modeling). The colorimetric substrate tests show that the functionality of our α-Galactosidase part is successful.
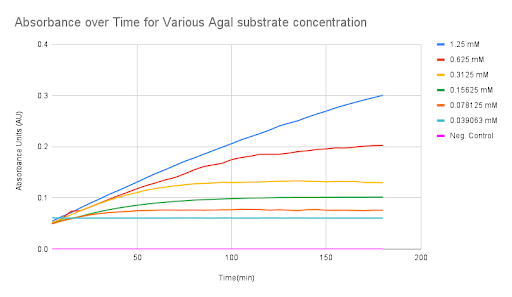
Figure 5. Absorbance over time for various α-Gal substrate concentrations
Mass Spectroscopy Trisaccharide Antigen Tests
To test a proof of concept functionality of α-Gal in cleaving the B-antigen trisaccharide and NAGA in cleaving the A-antigen trisaccharide, we carried out a reaction of the enzyme and its corresponding trisaccharide dissolved in 1X GlycoBuffer (New England BioLabs), deionized water, and purified BSA for 1 hour at 37°C. A-antigen and B-antigen trisaccharides were kindly supplied by Professor Dr. Todd Lowary from the Institute of Biological Chemistry at Academic Sinica [5].
For each enzyme, a non-specific trisaccharide served as a negative control and the trisaccharide specific to the enzyme served as the experimental unit (Table 1).
Table 1. Experimental setup for enzyme-antigen tests.

After the reaction, the reaction solution was passed through C18 columns to minimize impurities from the reaction solution. We then evaluated the flow through solution with a mass spectrometer to measure peaks in molar mass in order to determine the compounds present in the reaction solution. The original molar mass of the A-antigen is 713.35g/mol; therefore, after cleavage, two fragments with molar masses of 510.27g/mol and 203.08g/mol are expected to form. The original molar mass of the B-antigen is 672.32g/mol; therefore, after cleavage, two fragments with molar masses of 510.27g/mol and 162.05g/mol are expected to form (Fig 6).
References
1. Rahfeld, Peter, and Stephen G. Withers. “Toward Universal Donor Blood: Enzymatic Conversion of A and B to O Type.” Journal of Biological Chemistry, vol. 295, no. 2, Jan. 2020, pp. 325–34. DOI.org (Crossref), https://doi.org/10.1074/jbc.REV119.008164.
2. XTractorTM Buffer & xTractor Buffer Kit User Manual. (n.d.). 10.
3. Held, Paul. “Kinetic Analysis of β-Galactosidase Activity Using the PowerWave™ HT and Gen5™ Data Analysis Software .” BioTek, 16 Feb. 2007.
4. Biolabs, New England. “Typical Reaction Conditions for α-N-Acetylgalactosaminidase (P0734).” New England Biolabs: Reagents for the Life Sciences Industry, https://international.neb.com/protocols/2013/01/10/typical-reaction-conditions-p0734.
5. Meloncelli, Peter J., and Todd L. Lowary. “Synthesis of Abo Histo-Blood Group Type I and II Antigens.” Carbohydrate Research, vol. 345, no. 16, 16 Sept. 2010, pp. 2305–2322., https://doi.org/10.1016/j.carres.2010.08.012.
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25INCOMPATIBLE WITH RFC[25]Illegal AgeI site found at 246
Illegal AgeI site found at 549 - 1000COMPATIBLE WITH RFC[1000]
| None |
