Part:BBa_K3728000
Tol2 transposase (Oryzias latipes) - TPase
ThisTol2 transposon system is highly used in zebrafish transgenesis. The transposase protein (TPase) is from the Medaka fish (Oryzias latipes) aka Japanese rice fish, which catalyzes the transposition of the Tol2 elements through cut-and-paste mechanism. The minimal transposable Tol2 sequence (mTol2) contains 200-bp left arm and 150-bp right arm[1]. Up to 11kb DNA insert between Tol2 sequence can be integrated into the genome of nearly all vertebrates including zebrafish, frog, chicken, mouse, and human [2].
ThisA further application in synthetic biology was demonstrated by Jun Ni, et. al.[3], in which the recombinant TPase protein is fully functional in HeLa cell line and Zebrafish germline cells. In addition, the TPase can be expressed under T7 promoter in E. coli BL21 and purified with N-terminal 6xHis tag. The transposase is active in vitro and mediated the integration of DNA fragments between plasmids with Tol2 elements.
CONSTRUCTION – TPase/pSB1C3
ThisOryzias latipes Tol2 transposase (TPase) gene sequence was taken from UniProt database (UniProtKB - Q9PVN3) and optimized based on E. coli codon preference. The TPase gene was designed with 6xHis tag and a GS linker at N-terminus and synthesized by Integrated DNA Technologies, Inc. (IDT). The part was checked by colony PCR and restriction enzymes and further confirmed by sequencing (Fig. 1).
Figure 1 | 6xHis-GS-TPase/pSB1C3 construct check. DNAs were run electrophoresis on 1% agarose gel with 1kb marker. (a) 4 colonies were subject to PCR with TPase-specific forward and reverse primers (PCR product size: ~2000 bp). (b) The DNAs were extracted and digested by EcoRI and BamHI (2893, 870 and 235 bps).
PROTEIN EXPRESSION, PURIFICATION & ANALYSIS
ThisHis-tagged Tol2 transposase (TPase) was assembled with a T7 promoter and expressed in a cell-free in vitro transcription-translation (TXTL) system using the bacterial cytoplasmic extracts prepared from IPTG-induced E. coli Rosetta 2(DE3) cells. The His-tagged proteins were further purified through Nickel column. The protein concentration was measured and analyzed on SDS-PAGE and Coomassie Blue Staining (Fig. 2). The protein was shown at around 70 kDa as the same size as the predicted TPase protein (664 a.a., 75 kDa). The Elution #4, #5 and #6 were collected and used for further studies.
Figure 2 | His-Tol2 transposase was expressed in TXTL and purified by Nickel column. 10 μg of protein lysates were analyzed by SDS-PAGE and Coomassie Blue Staining using 4–12% gradient gel (NuPAGE™, Thermo Fisher Scientific Inc.) Lane: (1) PageRuler™ Prestained Protein Ladder, (2) E. coli Rosetta 2(DE3) cell extracts (no DNA control), (3) total lysates in TXTL, (4) flow-through, (5) wash-through, (6) Elution #4, (7) Elution #5, (8) Elution #6, (9) Elution #7, (10) Elution #8, (11) Elution #9.
IN VITRO INTEGRATION ASSAY
ThisIn vitro integration assay was used by Jun Ni, et al. to characterize the activity of purified recombinant Tol2 transposase (TPase) and the transposition of Tol2 mobile element[4]. We prepared the purified TPase from TXTL (Fig. 2) and performed PCR to generate KanR, ldhp-GFP-Tr and ldhp-amilCP-Tr (expressing blue color) DNA fragments flanked by 200-bp right and 150-bp left arms of pTol2 (Part:BBa_K3728002). The mixtures of TPase, Tol2 mobile inserts and a target plasmid of pSB1C3 were incubated at 30°C for 2 hours. The resulting DNAs were cleaned up and subjected to transform E. coli DH5α competent cells. The colonies displaying kanamycin resistance, green fluorescence or blue color were counted as successful jumping to plasmids by active purified TPase. And the integration rate was calculated by comparing with chloramphenicol resistance or red colonies from pSB1C3 backbone carrying Part:BBa_J04450 part (i.e., RFP coding device).
ThisGFP/Tol2-integrated plasmid can transform E. coli to exhibit weak to strong green fluorescence in Fig. 3. Two plasmids of GFP-positive bacteria were extracted and checked by restriction enzymes. They are larger than pSB1C3 when single cut on the backbone by ApaLI (Fig. 4b). The schematic map of Fig. 4a showed the possible position of integration by a BamHI-cut on the insert and a ApaLI-cut on the backbone (Fig. 4c). The rate of successful integration was calculated by the ratio of numbers of KanR, GFP and BLUE colonies to CmR or RED colonies, respectively (Fig. 4d). The ratio was between 0.2% to 0.9%, of which data are consistent with the observation by Jun Ni, et al[5]. In sum, we can modify plasmid DNAs in vitro with an insert between Tol2 mobile elements (DONOR) and purified TPase enzymes (HELPER) from TXTL reaction.
- Figure 3 | E. coli colonies on Cm agar plates were transformed by the mixture of GFP/Tol2 and pSB1C3 with TPase or without TPase as a control.
- Figure 4 | Possible integration map and ratio. (a) Schematic maps showed the predicted integration sites. (b, c) pSB1C3::GFP/Tol2 Clone #1 (lane 1) and #2 (lane 2) or pSB1C3 as a control (lane 3) were cut by ApaLI on the backbone (b) or cut by ApaLI with a BamHI cut on the insert (c). DNA was analyzed by electrophoresis on 1% agarose gel with a 1kb marker. (d) The successful integration ratios are calculated by the numbers of colonies of pSB1C3::KanR/Tol2 on Kan agar plate divided by those of pSBC13 (CmR) on Cm agar plates or by the numbers of pSB1C3::GFP or pSB1C3::BLUE divided by colony numbers of pSB1C3 (RED) on Cm agar plates such as shown in Figure 3.
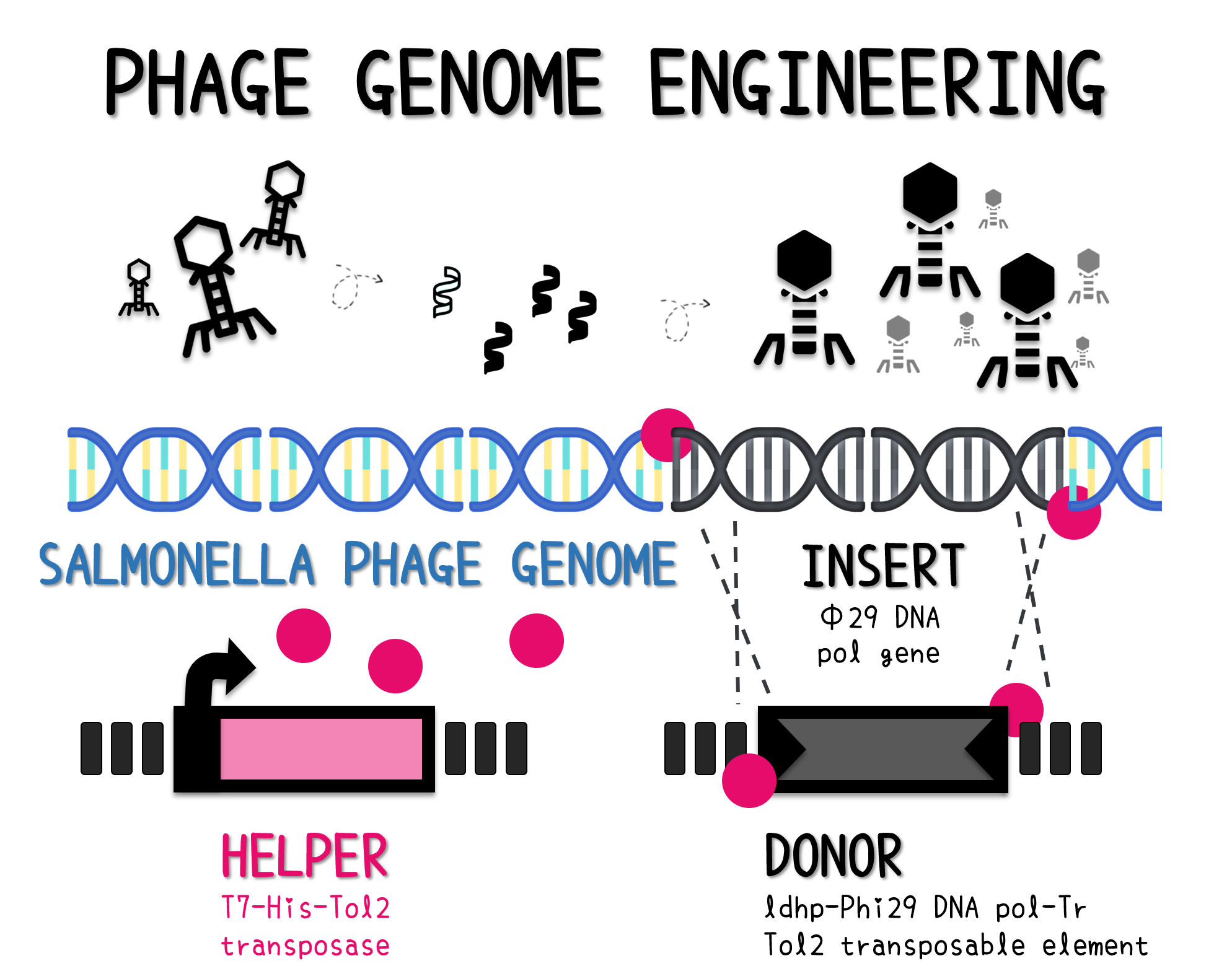
PHAGE ENGINEERING
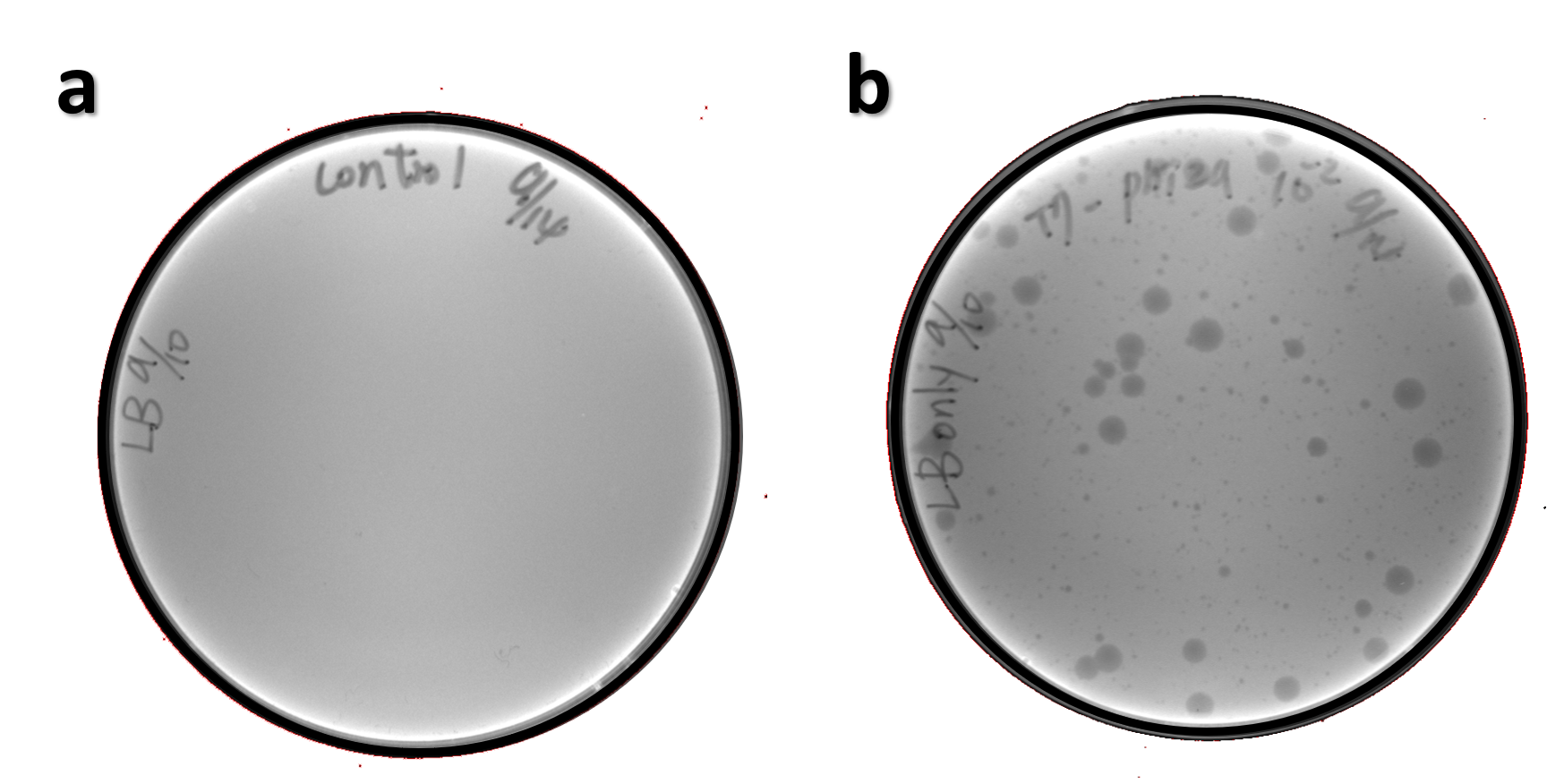
ThisThe genome of Salmonella phage #ST1 was extracted and engineered to carry phi29 DNA polymerase gene through the Tol2 transposon system. T7-His-Tol2 transposase was expressed in TXTL using IPTG-induced E. coli Rosetta 2(DE3) extracts, followed by purification with Nickel column. The DNA fragment of Tol2 transposable element carrying ldhp-Phi29 DNA polymerase-Tr (Ф29/Tol2) was amplified by PCR. The extracted Salmonella phage genome and the Ф29/Tol2 DNA fragment were incubated with Tol2 transposase at 30°C for 2 hours. The recombinant phage genome was subjected to TXTL based on the work of Jonghyeon Shin1, where T7 phage genome can be replicated, synthesized, and assembled in a single cell-free reaction. The infectious phage of Salmonella phage #ST1::Ф29 made by TXTL using Salmonella extracts was tested on plaque assay with a culture of Salmonella on the LB agar plate. As shown in Fig. 5b, visible different sizes of plaques were formed on the agar plate, demonstrating infectious phages were produced in our TXTL system. As a control, the phage gDNA without TXTL reaction displayed no plaques (Fig. 5a), indicating the live phages from TXTL are not from the contaminated DNA in the process of genomic DNA extraction.
Figure 5 | The recombinant phage synthesis in TXTL. Plaques were formed on a lawn of Salmonella culture on the LB agar plate from TXTL reaction (b) compared to no plaques without TXTL reaction (a).
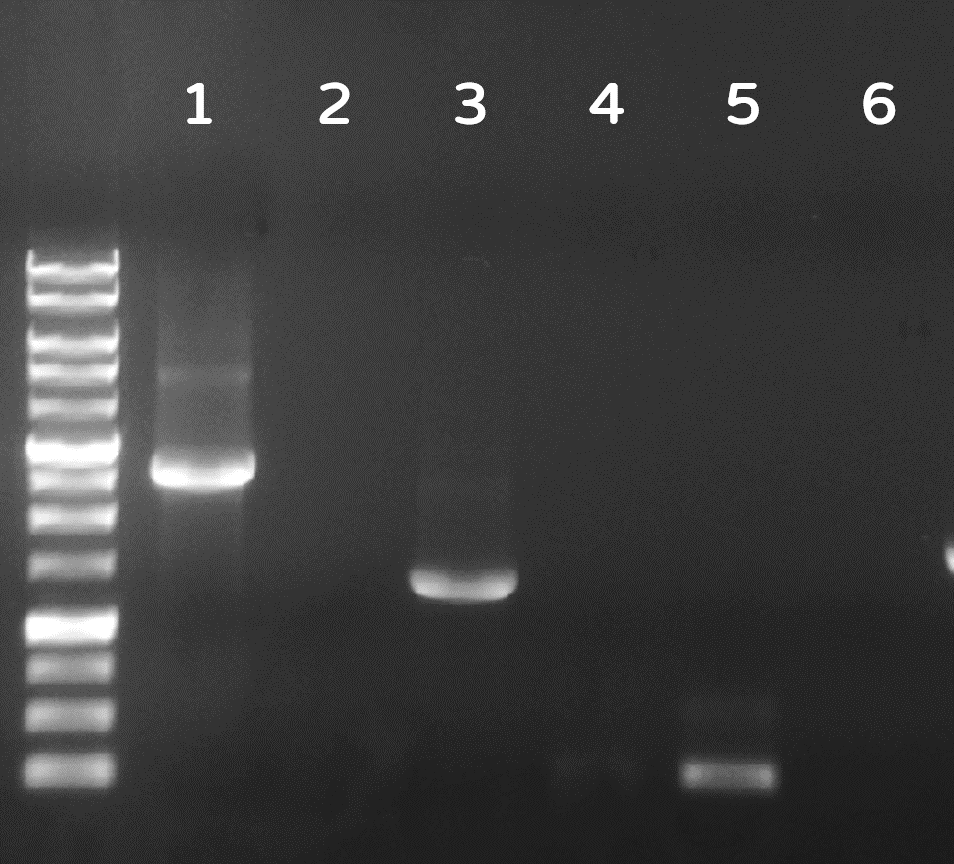
ThisDozens of plaques were screened by PCR with phi29 DNA polymerase gene-specific primers. A representative result on DNA gel electrophoresis was shown in Fig. 6, in which the successful Ф29/Tol2 insertion (Salmonella phage::Ф29 DNA polymerase, or #ST1::Ф29 for short) can be amplified by PCR with either Tol2 transposable element-specific primers or phi29 DNA polymerase-specific primers, compared to no PCR products from wild-type Salmonella phage #ST1, showing the success of our Salmonella phage engineering with phi29 DNA polymerase gene.
Figure 6 | Salmonella genome were checked by PCR with Tol2 transposable element-specific primers (lanes 1, 2) or with phi29 DNA polymerase gene-specific primer set 1 (lanes 3, 4) or set 2 (lane 5, 6). The odd numbers refer to Salmonella phage::Ф29 DNA polymerase (#ST1::Ф29), and the even numbers refer to wild-type Salmonella phage #ST1. The gel electrophoresis was performed on a 1% agarose gel with a 1kb DNA ladder.
APPLICATION - SALMONELLA DETECTION
Comparison to Commercial phi29 DNA polymerase
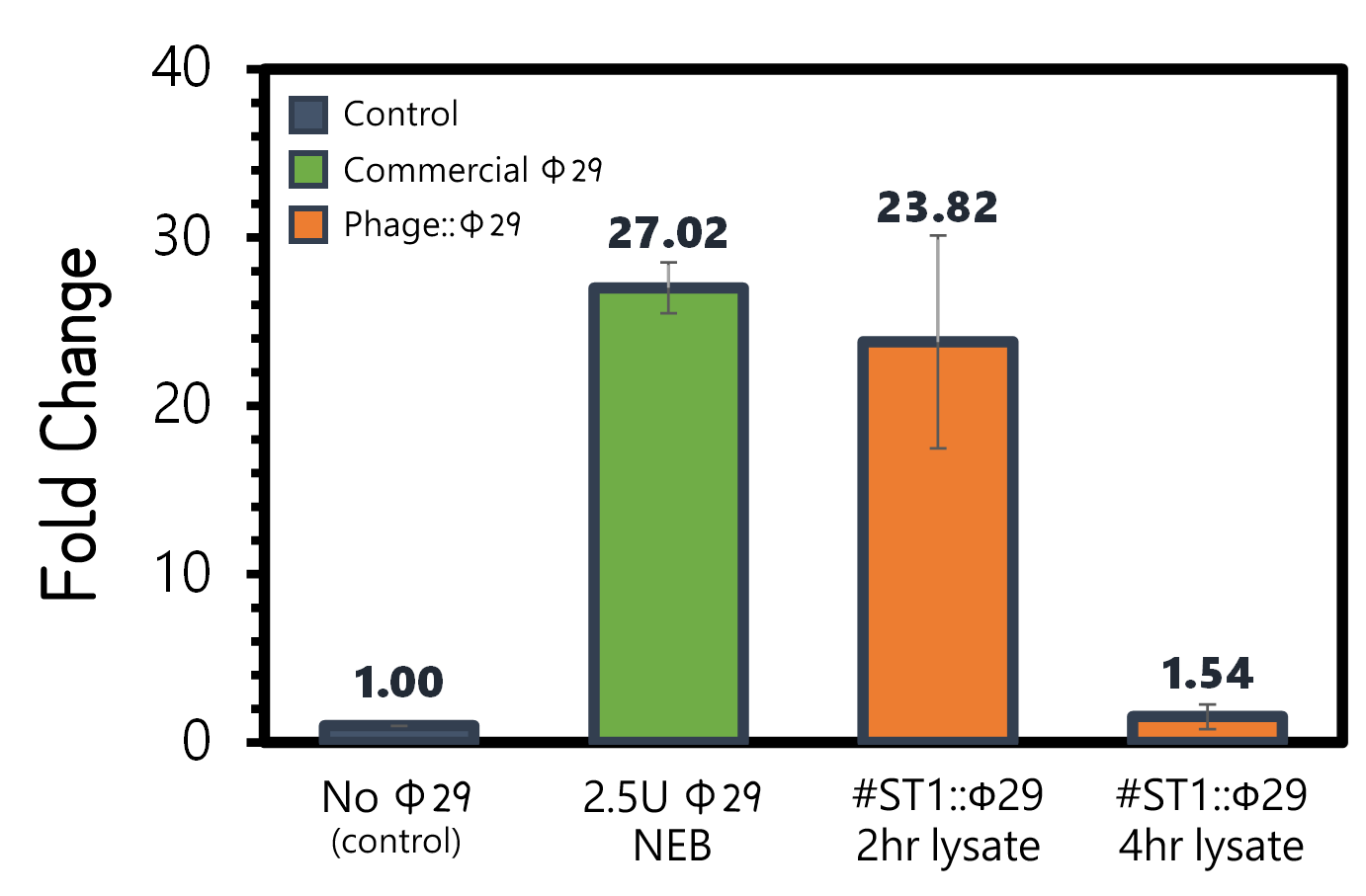
ThisAn overnight culture of Salmonella Typhimurium LT2 (~109 cells/ml) were infected by the engineered reporter phage #ST1::Ф29 at MOI=0.1 to produce phi29 DNA polymerase. The lysates were collected after 2 hr or 4 hr of treatment, and then subjected to RCA test. The lysate of phage-infected Salmonella at 2 hr can induce strong RCA reaction (24-fold change) comparable to 2.5U of a commercial phi29 DNA polymerase (NEB) in Fig. 7. Surprisingly, the lysate at 4 hr can not trigger any signal in RCA, suggesting a quick decay of phi29 DNA polymerase in the phage-infected bacterial lysates.
Figure 7 | RCA assay with NEB phi29 DNA polymerase or the Salmonella (109 cells/ml) lysates infected by #ST1::Ф29 at MOI=1 which were collected at 2 hr or 4 hr post infection. The fold changes were calculated by the fluorescence intensity of EvaGreen DNA binding of RCA materials without phi29 DNA polymerase as a background control. RCA was performed at 30°C for 1 hr.
Detection Time
ThisWe are wondering whether the time of bacterial lysis by phage infection affects the stability of phi29 DNA polymerase protein. An isolated phage may be featured by a latent time (phage generation time in a bacterial cell) and a burst size (numbers of phage produced per bacterial cell). And the latent time and burst size are in a relationship in terms of bacterial density[6] and MOI of phage infection[7].
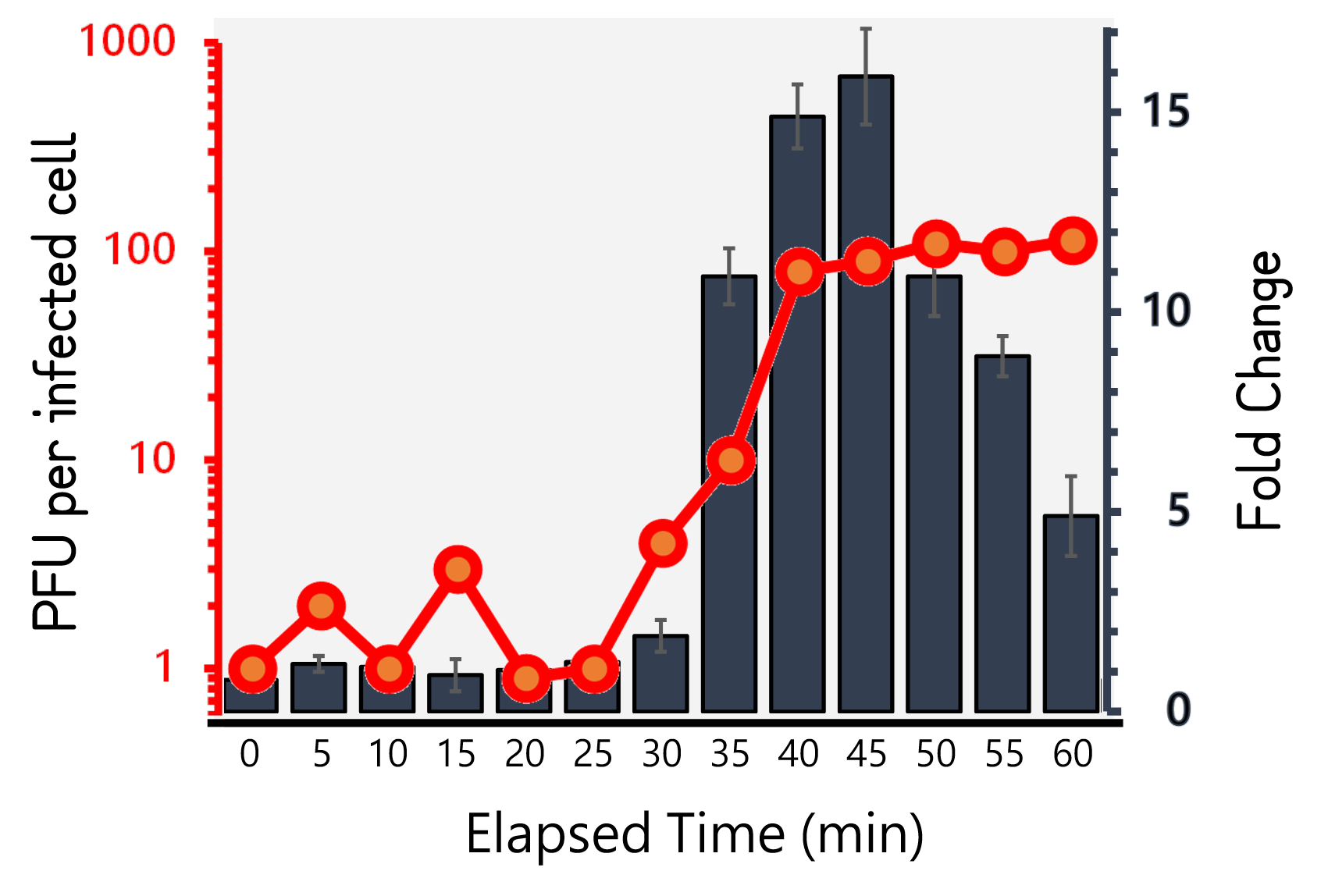
ThisTherefore, we performed the experiment to figure out the latent time and burst size of our Salmonella phage #ST1::Ф29 and the time course of RCA signals during phage infection in Salmonella. 105 cells/ml of Salmonella were infected by phage #ST1::Ф29 at MOI=1. The lysates collected by an interval of 5 min until 60 min and subjected to plaque assays and RCA test. As Fig. 8 demonstrated, the phages were released at around 40 min (latent time) to a plateau level with a burst size of average 98.4±14. Interestingly, the RCA signal increased dramatically at 35 min, achieved a high level around 40-45 min, and dropped significantly thereafter, that are consistent with our speculation of the correlation between phage lysis time (latent time) and phi29 DNA polymerase protein functionality.
Figure 8 | Salmonella phage #ST1::Ф29 latent time (min) and burst size (PFU per infected cell, the left Y axis) at MOI=1, and the relationship to RCA assay (fold change, the right Y axis). Phage-infected Salmonella lysates were harvested for 1 hour at an interval of 5 min. The lysates were subjected to plaque assays and RCA reaction followed by stained with EvaGreen dye. The burst sizes were counted by numbers of plaques. The RCA signal were read at Ex/Em=500/530 nm and divided by the background level without phi29 DNA polymerase.
Detection Limit with Our Hardware
ThisImagine an application of real Salmonella diagnosis in a food or drink. A contaminated sample collected in a large volume may find 1-100 CFU/ml of bacteria[8]. Large volume and bacterial density are key parameters to affect the result of Salmonella detection with our product.
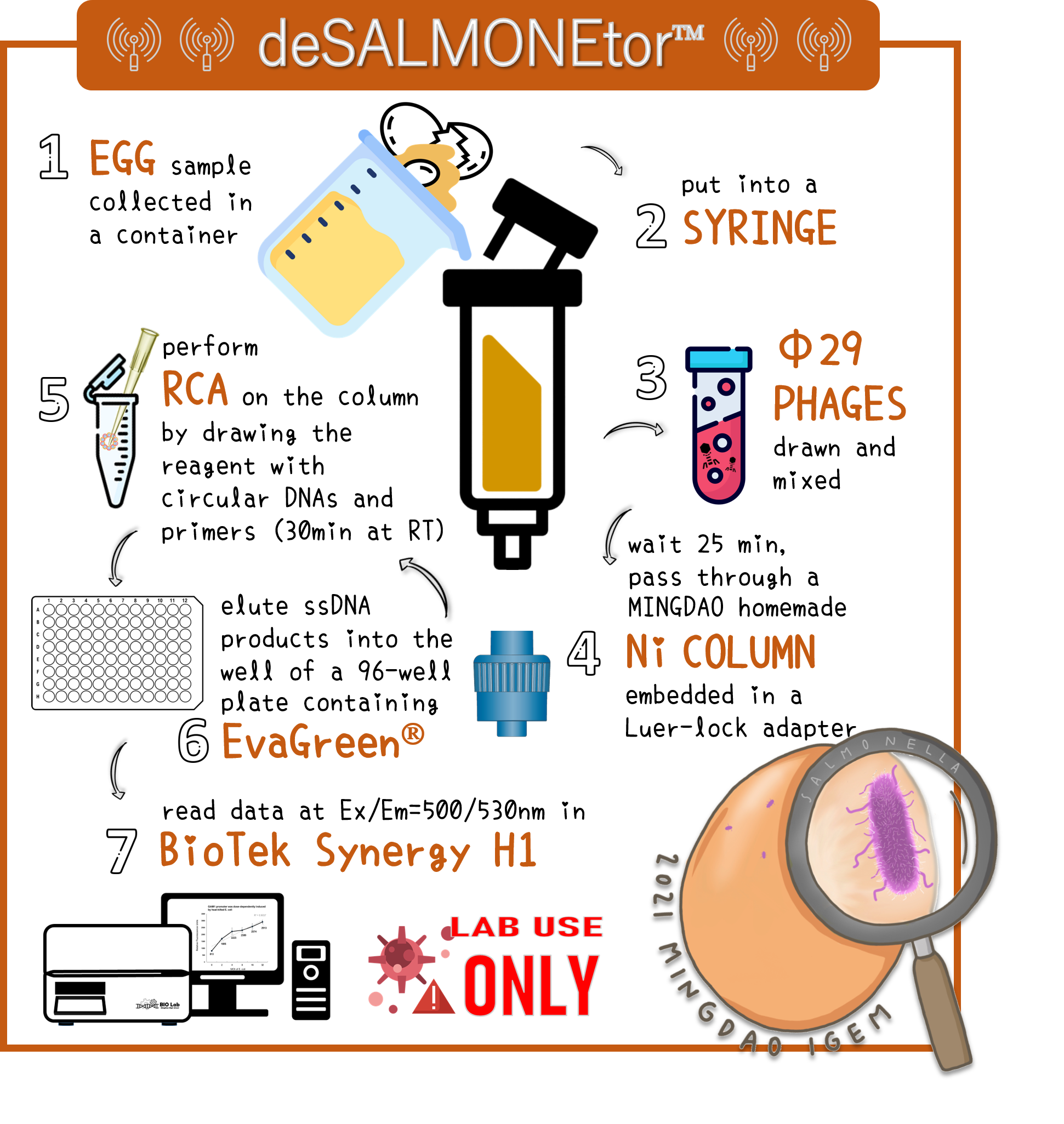
ThisTo overcome the large volume of a sample, we were inspired by Nickel column purification, in which His-tagged phi29 DNA polymerase were bound. Therefore, we think this method may enrich the His-phi29 DNA polymerase from the sample of large volume. Go to our page of (HARDWARE) for such a design. And the schematic diagram was shown here.
THE LIMIT OF SALMONELLA PHAGE REPORTER
COMPARISON WITH CURRENT METHODS
USAGE: SAFETY ISSUE - GENE DRIVE
Reference
- ↑ Urasaki A, Morvan G, Kawakami K. Functional dissection of the Tol2 transposable element identified the minimal cis-sequence and a highly repetitive sequence in the subterminal region essential for transposition. Genetics. 2006 Oct;174(2):639-49. doi: 10.1534/genetics.106.060244.
- ↑ Kawakami K. Tol2: a versatile gene transfer vector in vertebrates. Genome Biol. 2007;8 Suppl 1(Suppl 1):S7. doi: 10.1186/gb-2007-8-s1-s7
- ↑ Ni J, Wangensteen KJ, Nelsen D, Balciunas D, Skuster KJ, Urban MD, Ekker SC. Active recombinant Tol2 transposase for gene transfer and gene discovery applications. Mob DNA. 2016 Mar 31;7:6. doi: 10.1186/s13100-016-0062-z.
- ↑ Ni J, Wangensteen KJ, Nelsen D, Balciunas D, Skuster KJ, Urban MD, Ekker SC. Active recombinant Tol2 transposase for gene transfer and gene discovery applications. Mob DNA. 2016 Mar 31;7:6. doi: 10.1186/s13100-016-0062-z
- ↑ Ni J, Wangensteen KJ, Nelsen D, Balciunas D, Skuster KJ, Urban MD, Ekker SC. Active recombinant Tol2 transposase for gene transfer and gene discovery applications. Mob DNA. 2016 Mar 31;7:6. doi: 10.1186/s13100-016-0062-z
- ↑ Abedon ST. Selection for bacteriophage latent period length by bacterial density: A theoretical examination. Microb Ecol. 1989 Sep;18(2):79-88. doi: 10.1007/BF02030117.
- ↑ Atel, IR, and K. K. Rao. Bacteriophage burst size as a function of multiplicity of infection. Current Science 1984 53(4): 198–200.
- ↑ Gwimbi P, George M, Ramphalile M. Bacterial contamination of drinking water sources in rural villages of Mohale Basin, Lesotho: exposures through neighbourhood sanitation and hygiene practices. Environ Health Prev Med. 2019 May 15;24(1):33. doi: 10.1186/s12199-019-0790-z.
Note: The map was generated and sponsored by SnapGene.
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21INCOMPATIBLE WITH RFC[21]Illegal BamHI site found at 1554
- 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000COMPATIBLE WITH RFC[1000]
| None |