Part:BBa_K3578010
AnGlu Starch degradation
EXP, AnGlu, XPR2t part will be digested by BsaI and then assembled as the module 1 through the T4 ligase. The module 1 and Leu selection marker will be digested by SapI and then assembled as “EXP-AnGlu-XPR2t-Leu” expression cassette through the T4 ligase. The cassette can be transformed into the Yarrowia lipolytica plotoplast and integrated into the genome for AnGlu efficient expression.
Sequence and Features
- 10INCOMPATIBLE WITH RFC[10]Illegal EcoRI site found at 3683
Illegal EcoRI site found at 3733
Illegal EcoRI site found at 4695
Illegal SpeI site found at 756 - 12INCOMPATIBLE WITH RFC[12]Illegal EcoRI site found at 3683
Illegal EcoRI site found at 3733
Illegal EcoRI site found at 4695
Illegal NheI site found at 817
Illegal SpeI site found at 756 - 21INCOMPATIBLE WITH RFC[21]Illegal EcoRI site found at 3683
Illegal EcoRI site found at 3733
Illegal EcoRI site found at 4695
Illegal BglII site found at 2537
Illegal BglII site found at 5964
Illegal BamHI site found at 3157
Illegal XhoI site found at 4678
Illegal XhoI site found at 4707 - 23INCOMPATIBLE WITH RFC[23]Illegal EcoRI site found at 3683
Illegal EcoRI site found at 3733
Illegal EcoRI site found at 4695
Illegal SpeI site found at 756 - 25INCOMPATIBLE WITH RFC[25]Illegal EcoRI site found at 3683
Illegal EcoRI site found at 3733
Illegal EcoRI site found at 4695
Illegal SpeI site found at 756
Illegal NgoMIV site found at 3821
Illegal AgeI site found at 3364
Illegal AgeI site found at 5292 - 1000COMPATIBLE WITH RFC[1000]
Usage and Biology
Yarrowia lipolytica can use starch as sole carbon sources when the AnGlu is expressed successfully, which means the AnGlu has the ability to degrade the starch efficiently.
Results
1.Plasmid Construction
We constructed the plasmids according to DESIGN page. There are four parts in our plasmid library: a promoter-EXP (BBa_K3578000), a terminator-XPR2t (BBa_K3578002), expression genes glucoamylase (BBa_K3578001), and the Leu selection marker (BBa_K3578004). By BsaI and SapI IIs enzymes digestion and T4 ligase, we successfully constructed the plasmids which separately carried the AnGlu expression cassette with Leu selection marker (Figure 1). Then plasmids were linearizd with EcoR I and transformed into Y. lipolytica plotoplast. The AnGlu expression cassette with Leu selection marker DNA fragment will be randomly integrated into genome loci.
By the PCR experiment, we randomly selected 10 strains in which AnGlu and Leu expression cassette have been successfully integrated into the genome. We designed the primers(F/R) to identify the Anglu expression cassette. The primer design was shown in the figure 2, and the predicted PCR result is 2736 bp. The electrophoresis results showed that about 3000 bp bands were obtained by using Anglu expression cassette as template, the electrophoresis results were consistent with our expectation(Figure 2).
2.The expression of AnGlu in Y. lipolytica is sufficient to allow growth on starch
In order to verify whether the AnGlu possess the ability for starch degradation. Engineered Y. lipolytica expressing the AnGlu (Polg-AnGlu) individually were cultured in the starch medium in which the 30 g/L starch was used as the sole carton source. In addition, the strain Po1g was also cultured in the starch medium with the addition of 0.4g/L leucine as the control (Polg-Leu). The OD was measured every 24 hours. As shown in Figure 3, Po1g-AnGlu strain can grow well in the starch medium, it means that AnGlu had the relatively higher starch utilization efficiency. The result can also be directly observed as shown in Figure 4, in which Polg-AnGlu possess a much higher cell density. In summary, the expression of AnGlu in Y. lipolytica is sufficient to allow growth on starch.
3.High C/N ratio is more conducive to oil accumulation
Since the Polg-AnGlu strain can growth well on the starch medium, we then measured the lipid titer and content. The Polg-AnGlu can produce up to 0.76 ± 0.04 g/L lipid and 14.66 ± 0.43% of DCW as fatty acids in the starch media and a C/N ratio of 30. The titer and content were relatively low. Previous researchers proved that higher C/N ratio is more conducive to lipid accumulation when glucose, xylose and glycerin were used as the carbon source. In order to verify whether the higher C/N ratio in the starch media also help improve the lipid biosynthesis capability, we regulated the C/N ratio by adding different concentrations of starch (Figure 5). The C/N ratio was 30, 60, 90. These statistics showed that when C/N ratio was increased to 60, the lipid titer increased from 0.76 ± 0.04 g/L to 1.07 ± 0.02 g/L, a 55% improvement. when C/N ratio was increased to 90, the lipid titer further increased to 1.17 ± 0.02 g/L. Similar results about the increasing lipid content were also observed. The lipid content increased from 14.66 ± 0.43% to 24.06 ± 0.13% of DCW, a 64% improvement. In summary, it shows that improving C/N ratio is an effective strategy to strengthen lipid accumulation.
Finally, in order to verify the above conclusion, lipid bodies visualization was carried out by addition of 1 mg/mL Nile red to the cell suspension. The results showed that the color of lipid bodies is brighter and the size is relatively bigger when C/N ratio was controlled as 90 rather than 30. The above results indicate that the ratio of C/N is closely related to the lipid accumulation (Figure 6).
Improve From NNU-China 2021
Group:iGEM Team NNU-China 2021
Author: Yan Xu
Background
Herein, to improve the production of AMPs, the RBS library of T7 RNA polymerase based on Escherichia coli BL21 (DE3) was constructed, and then the most suitable expression host could be screened by high throughput screening. Besides, promoter engineering is also an effective strategy to improve the expression of genes. Promoter plays a significant role in gene transcription because it controls the binding of RNA polymerase to DNA.
Therefore, we are trying to use promoter engineering to improve BioBrick BBa_K3578010 from the 2020 iGEM worldshaper-Nanjing Team, and improve the ability of Yarrowia lipolytica to utilize the starch as an improvement.
We hypothesized that the expression level of glucoamylase could be increased through using the more potent promoters. For this, we scanned the genome of Y. lipolytica and characterized 20 endogenous promoters in Y. lipolytica.
Design
Given Y. lipolytica's metabolism with a high propensity for flux to tricarboxylic acid (TCA) cycle intermediates, the promoters chosen are primarily centering around the carbon metabolism and tricarboxylic acid cycle. We selected 20 endogenous promoters (Table1) in Y. lipolytica for genetic parts characterization. Specifically, these genes include YALI0E00638g, YALI0F07711p, YALI0A15972p, YALI0E26004p, YALI0D02277p, YALI0F16819p, YALI0D06325p, YALI0D23683p, YALI0F02607p, YALI0D06215p, YALI0C19965p, YALI0E02090p, YALI0D06930p, YALI0A15147p, YALI0A16379g, YALI0F0960p, tYALI0C19624p, YALI0B10406p, YALI0E18590p, and YALI0D17864p.
Next, to test the strength of these promoters, we amplified the 1500 bp promoter and 5′ UTR (5′-untranslated region) sequences of the target genes, and further constructed 20 recombinant plasmids by Gibson Assembly method. Recombinant plasmids were constructed based on the original plasmid pYLXP'-PTEF-Nluc harboring with the nanoluc luciferase gene (Nluc) as the reporter by Gibson Assembly method. For example, the recombinant plasmid of pYLXP’-P1-Nluc was constructed using linearized pYLXP'-PTEF-Nluc (digested by AvrII and NheI) and the PCR-amplified promoter P1 sequences by Gibson Assembly.
Then, 20 recombinant plasmids were transformed into Y. lipolytica po1fk. In brief, one milliliter cells were harvested during the exponential growth phase (16-24 h) from 2 mL YPD medium (yeast extract 10 g/L, peptone 20 g/L, and glucose 20 g/L) in the 14-mL shake tube, and washed twice with 100 mM phosphate buffer (pH 7.0). Then, cells were resuspended in 105 µL transformation solution, containing 90 µL 50% PEG4000, 5 µL lithium acetate (2M), 5 µL boiled single stand DNA (salmon sperm, denatured) and 5 µL DNA products (including 200-500 ng of plasmids, lined plasmids or DNA fragments), and incubated at 39 ℃ for 1 h, then spread on selected plates. The transformants were selected by using the leucine-defective solid medium.
Subsequently, shaking flasks were performed to characterize the strength of promoters by the Nluc analysis, which has been reported in the previous study (Liu et al., 2019).
Furthermore, the identified more potent promoter was used to express glucoamylase.

- Table 1: The genetic information of the selected 20 endogenous promoters
Result
After amplifying the 1500 bp promoter and 5′ UTR (5′-untranslated region) sequences of the target genes, we assembled these sequences with linearized pYLXP'-PTEF-Nluc (digested by AvrII and NheI) to construct the recombinant plasmids (Fig. 1 and Fig. 2), including pYLXP’-P1-Nluc, pYLXP’-P2-Nluc, pYLXP’-P3-Nluc, pYLXP’-P4-Nluc, pYLXP’-P5-Nluc, pYLXP’-P6-Nluc, pYLXP’-P7-Nluc, pYLXP’-P8-Nluc, pYLXP’-P9-Nluc, pYLXP’-P10-Nluc, pYLXP’-P11-Nluc, pYLXP’-P12-Nluc, pYLXP’-P13-Nluc, pYLXP’-P14-Nluc, pYLXP’-P15-Nluc, pYLXP’-P16-Nluc, pYLXP’-P17-Nluc, pYLXP’-P18-Nluc, pYLXP’-P19-Nluc, and pYLXP’-P20-Nluc.

- Fig. 1 The maps of recombinant plasmids for characterizing the strength of promoters
These recombinant plasmids were further transformed into Y. lipolytica po1f (Fig. 2), and obtained strains po1f-P1-Nluc, po1f-P2-Nluc, po1f-P3-Nluc, po1f-P4-Nluc, po1f-P5-Nluc, po1f-P6-Nluc, po1f-P7-Nluc, po1f-P8-Nluc, po1f-P9-Nluc, po1f-P10-Nluc, po1f-P11-Nluc, po1f-P12-Nluc, po1f-P13-Nluc, po1f-P14-Nluc, po1f-P15-Nluc, po1f-P16-Nluc, po1f-P17-Nluc, po1f-P18-Nluc, po1f-P19-Nluc, and po1f-P20-Nluc. Next, we performed the shaking flask of these engineering strains and tested the Nluc analysis.

- Fig. 2 A. The processes of constructing plasmids. B. Characterizing the strength of promoters
The results of the Nluc analysis showed that strains po1f-P2-Nluc and po1f-P17-Nluc generated the highest Nluc activities compared with other strains (Fig. 3), indicating promoter P2 and P17 are two strong promoters.

- Fig. 3 Nanoluc luciferase gene was used as a reporter gene to quantify the promoter strength.
Then, we replaced the promoter of plasmid pUC-Pexp-AnGlu(BBa_K3578010)with identified promoters P2 and P17, and obtained the recombinant plasmids pUC-P2-AnGlu and pUC-P17-AnGlu (Fig. 4), respectively. Further, we transformed plasmids pUC-P2-AnGlu and pUC-P17-AnGlu into po1f, and obtained strains po1f-P2-AnGlu and po1f-P17-AnGlu. Subsequently, we performed the shaking flask with using starch as the sole carbon source.

- Fig. 4 The maps of recombinant plasmids of pUC-P2-AnGlu and pUC-P17-AnGlu
As shown in Fig. 5, we can see that po1f-P2-AnGlu and po1f-P17-AnGlu both showed better cell growth than that of the control strain po1f-Pexp-AnGlu (BBa_K3578010), indicating that glycolase has been overexpressed by promoters P2 and P17. Moreover, among these strains, the cell growth of po1f-P2-AnGlu was the highest. That is to say that the use of strong promoter expression can improve glycolase activity, indicating that our strategy is indeed effective.
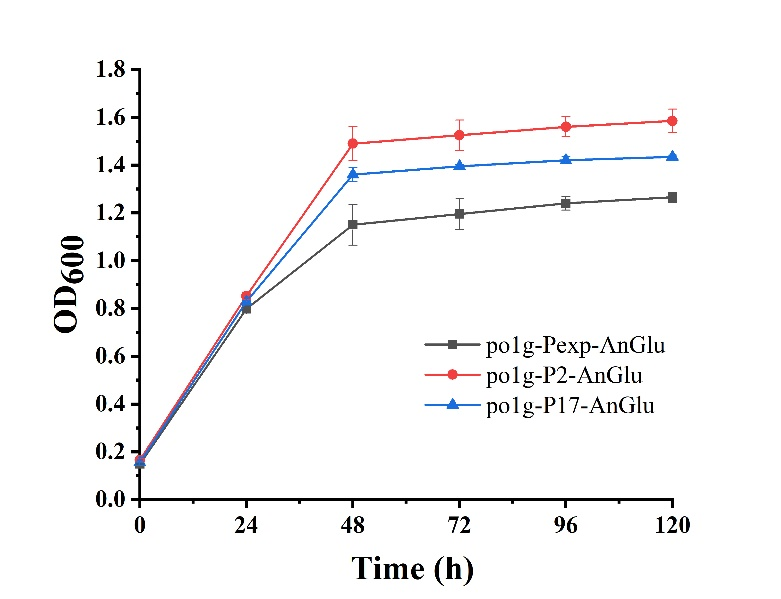
- Fig. 5 The maps of recombinant plasmids of pUC-P2-AnGlu and pUC-P17-AnGlu
References
Liu, H., Marsafari, M., Deng, L., Xu, P. 2019. Understanding lipogenesis by dynamically profiling transcriptional activity of lipogenic promoters in Yarrowia lipolytica. Appl Microbiol Biotechnol, 103(7), 3167-3179.
| None |






