Part:BBa_K3832000
aroG (Mutant S211F)
This part encodes a mutant of AroG (3-deoxy-7-phosphoheptulonate synthase, EC 2.5.1.54 ), in which serine at 211 was replaced by phenylalanine. The mutant can relieve the allosteric inhibition of phenylalanine, thus increasing the catalytic rate and downstream product yield.
The enzyme that encoded by the sequence catalyze the following reactions: phosphoenolpyruvate+D-erythrose 4-phosphate+H2O =3-deoxy-D-arabino-hept-2-ulosonate 7-phosphate +phosphate
Derived from E.coli DH5alpha strain.
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21INCOMPATIBLE WITH RFC[21]Illegal XhoI site found at 934
- 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000COMPATIBLE WITH RFC[1000]
1.Useage & Biology
This part encodes a mutant of aroG(as the improvement of part (BBa_K1060000). It can be expressed in Prokaryote cells, and functionally verified in E.coli DH5alpha strain.
Under physiological conditions, aroG catalyze the reaction phosphoenolpyruvate + D-erythrose 4-phosphate + H2O = 3-deoxy-D-arabino-hept-2-ulosonate 7-phosphate + phosphate. This reaction is a key branching point of the glycolysis and shikimate pathways. Expression of aroG can lead to more substrate into the shikimate pathway, which can improve the yield of downstream products as tryptophan, phenylalanine, tyrosine and benzazole.
In our project, aroG-s211f is used to improve the production of tryptophan. Considering the over-expression of aroG-s211f could significantly reduce the amount of substrate (glucose) entering the glycolysis reaction, in turn affects the normal process of cell proliferation, the expression of aroG is designed under strict control by Toggle-switch circuit (View our design on (Team:XJTU-China/Design - 2021.igem.org)).
2.Structure
3.Characterization & Measurement
We have constructed the inducible circuit (BBa_K3832008)to characterize and measure the function of aroG-s211f, by detecting the yield of tryptophan. Besides, to verify the impact of aroG-s211f to the cell proliferation, we measured the growth curve of the wild-type E.coli and the engineered E.coli with aroG-s211f.
Fig. 3.1 shows the yield of tryptophan. The engineered E.coli with aroG-s211f induced by 1mM IPTG resents a increased production of tryptophan, comparing in absent of IPTG or aroG-s211f (wild-type E.coli).
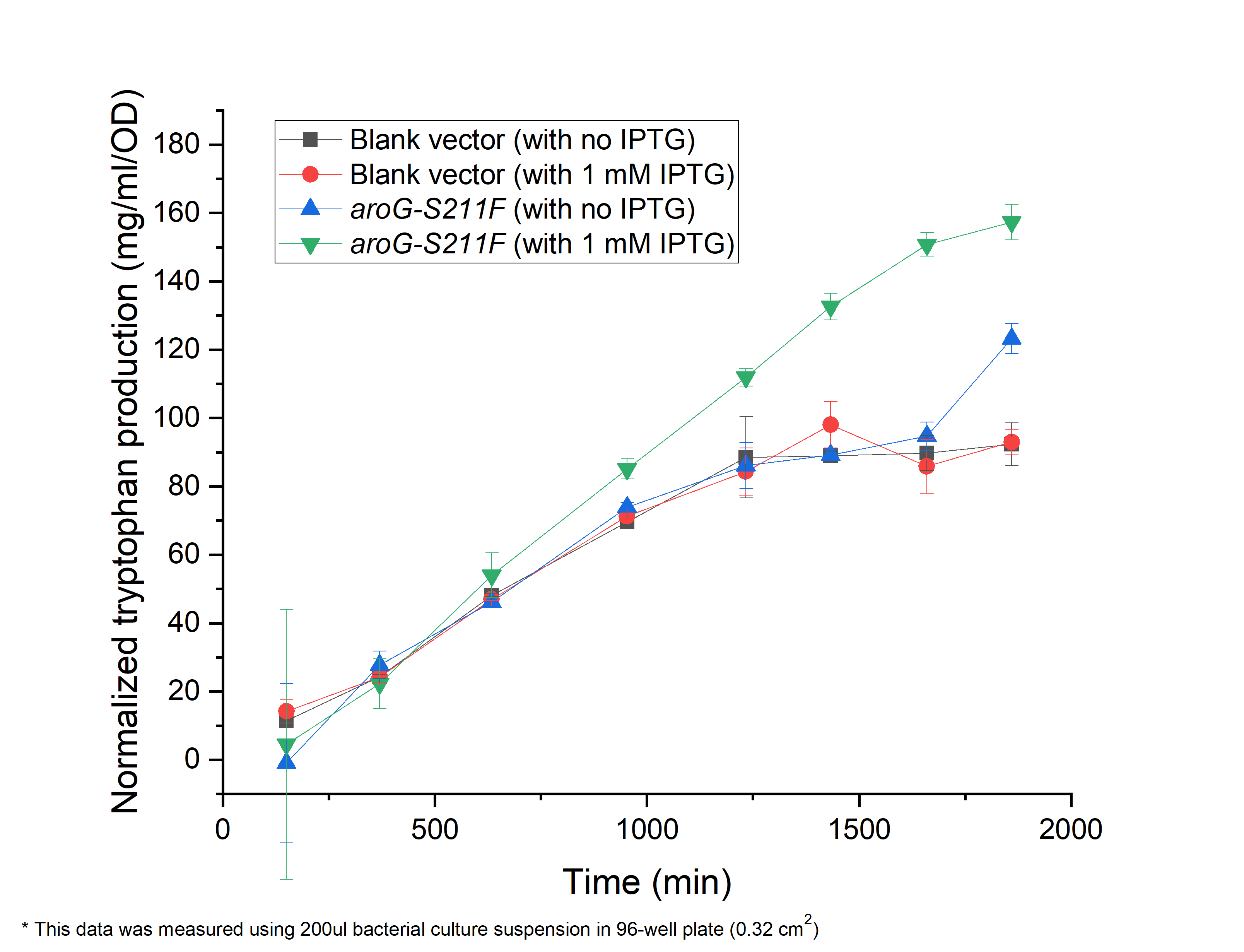 Fig. 3.1 The relationship between tryptophan concentration in culture medium and culture time. The concentration of tryptophan is measured by PDAB chromogenic method. The vertical axis shows the absorbance under 590nm, reflecting the relevant concentration of tryptophan.
Fig. 3.1 The relationship between tryptophan concentration in culture medium and culture time. The concentration of tryptophan is measured by PDAB chromogenic method. The vertical axis shows the absorbance under 590nm, reflecting the relevant concentration of tryptophan.
As is showed in Fig. 3.2, by using the Logistic equation to fit the growth curve, the inhibitory effect of aroG expression on cell proliferation was verified (as the parameter r decreased in E.coli with aroG-s211f and induced by IPTG, which represents the reciprocal of the time it takes for the population to double).
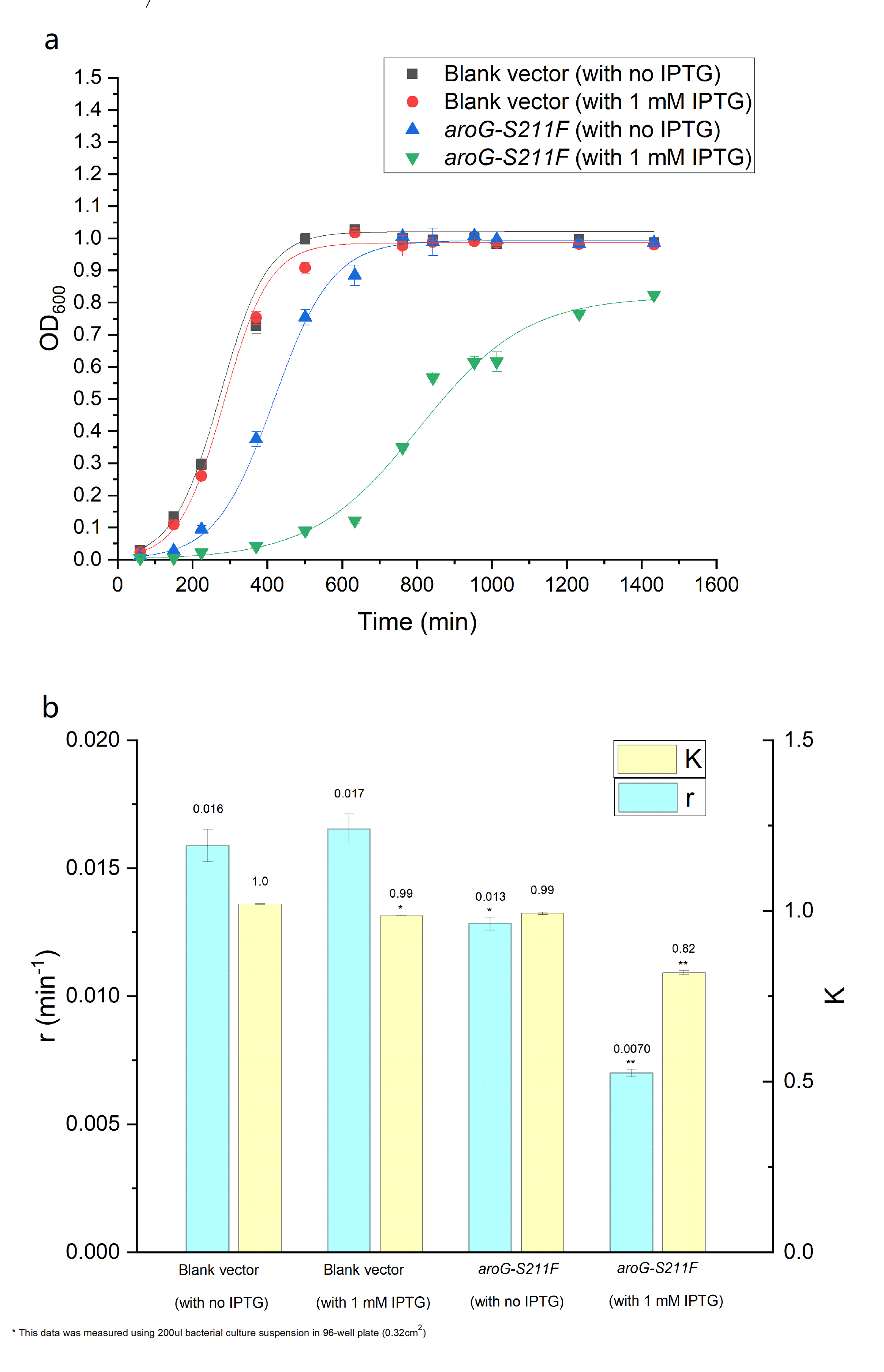 Fig. 3.2 (a) The population density of E.coli was measured at 600nm by colorimetry. The scatter represents the result of the measurement. The Logistic equation was used to fit the growth curve, and the fitting results were shown in the curve. (b) and (c) respectively show the growth parameters K (environmental capacity) and r (intrinsic growth rate) of different experimental groups obtained from the fitting results in (a).
Fig. 3.2 (a) The population density of E.coli was measured at 600nm by colorimetry. The scatter represents the result of the measurement. The Logistic equation was used to fit the growth curve, and the fitting results were shown in the curve. (b) and (c) respectively show the growth parameters K (environmental capacity) and r (intrinsic growth rate) of different experimental groups obtained from the fitting results in (a).
Here wild-type DH5alpha has been used as the control, which can represent the function and character of wild-type aroG(BBa_K1060000), for this gene is contained in the genome of E.coli.
4.Modeling
| biology | E.coli |
| kegg | EC 2.5.1.54 |
| protein | aroG |
