Part:BBa_K3331005
nodA(MazF)
ndoA (ydcD, mazF) [ Bacillus subtilis subsp. subtilis str. 168 ]
Encodes EndoA, an endoribonuclease that inactivates cellular mRNAs by cleaving them at specific, but frequently occurring sites. EndoA has similar cleavage specificity to MazF and PemK and yields cleavage products with 3′‐phosphate and 5′‐hydroxyl groups, typical of EDTA‐resistant degradative RNases.
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000COMPATIBLE WITH RFC[1000]
Characterization
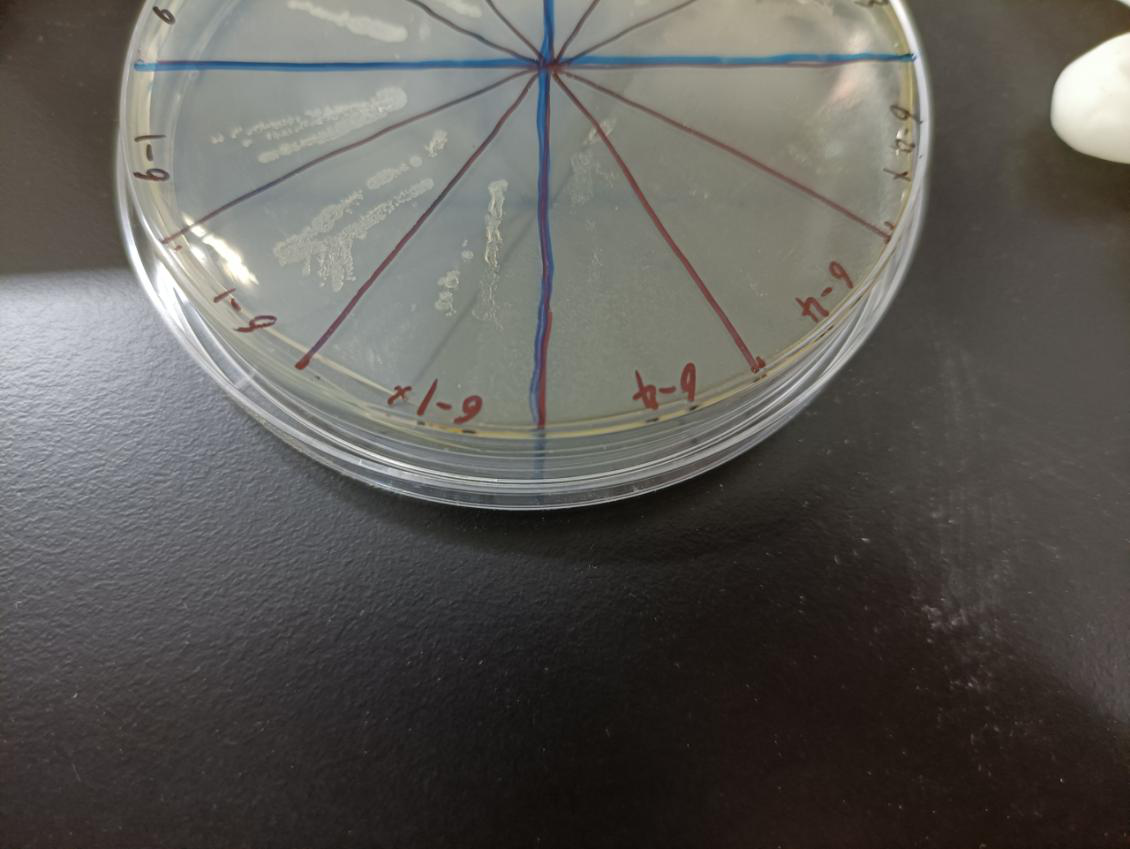
In the figure, the bacteria in this plate contained plasmids of pBAD-MazF. Three of 6-1 groups were the control group without arabinose induction, and three of 6-4 groups were the experimental group with 1.5 M/L arabinose induction. Where, the dilution factor of the group with * is 10^4, and the dilution factor of the group without * is 10^6.
Result: The bacteria without arabinose induction grow normally, while with arabinose induction, the bacteria suicide.
| None |