Part:BBa_K3617007
TMDWsc1-TEV_recseq-LexA-VP16
The biobrick is a membrane bound transcription factor designed to activate transcription of a reporter in Saccharomyces cerevisiae after it is cleaved by a TEV protease. The transcription factor is described in BBa_K165020; it is composed of the lexA DNA binding domain from Escherichia coli and an activation domain from the herpes simplex virus VP16. As opposed to BBa_K165020, this biobrick is localized at the plasma membrane through a flexible linker and a transmembrane domain. The linker includes a cut site for the TEV protease and upon cleavage the transcription factor is transported to the nucleus where it activates transcription.
Usage
The biobrick is designed to function as an accessory protein in S. cerevisiae biosensors for interleukins. It is expressed together with two modular receptor proteins also located at the plasma membrane. The receptors have human interleukin receptors extracellularly and either the C- or N-terminal part of a split TEV protease intracellularly. Split TEV protease is a TEV protease divided into two halves, which only regains its proteolytic activity when the halves are complemented. When the extracellular parts of the interleukin receptor proteins bind to the target interleukin, the receptors come together causing the split TEV protease to be complemented and regain proteolytic activity. One split TEV protease can then cleave multiple copies of the biobrick leading to amplification as well as transduction of the signal to the nucleus.
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21INCOMPATIBLE WITH RFC[21]Illegal BamHI site found at 507
- 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000COMPATIBLE WITH RFC[1000]
This biobrick consists of multiple parts; An endoplasmatic reticulum import signal peptide from the Saccharomyces cerevisiae cell wall integrity and stress response component 1 (Wsc1) receptor in S. cerevisiae, the transmembrane receptor of Wsc1, and the transcriptional activation domain of VP16 from herpes simplex virus, which is linked the LexA DNA binding domain from Escherichia coli. The transmembrane domain and the synthetic transcription factor are separated by a flexible GS linker followed by a seven amino acid consensus sequence (ENLYFQâG) for the cut site for Tobacco Etch Virus Nuclear Inclusion protein a (TEV protease).
Sequence optimization
The sequence was codon optimized for S. cerevisiae. The recognition sequences for SpeI, XbaI, NotI, EcoRI, PstI were avoided to follow the RFC10 standard.
Structure and function
The signal peptide included in the biobrick, and the transmembrane domain are endogenous to S. cerevisiae and stem from the cell wall integrity sensor Wsc1. They are used in yeast-two-hybrid assays of membrane protein-protein interactions to increase the chance of localization to the plasma membrane as described by Ivanusic et al. In the biobrick they cause the protein to be localized at the plasma membrane.
Between the transmembrane domain and the transcription factor is a flexible linker with a cut site for the TEV protease. Cleavage of the linker causes the transcription factor to localize to the nucleus, where it activates the transcription of the gene downstream from its promoter.
The LexA DNA binding domain of the synthetic transcription factor binds to the lexAop which is incorporated into an synthetic promoter consisting of six lexAop sequences and the core promoter from the eno1 promoter endogenous to S. cerevisiae. Since the transcription factor does not regulate endogenous gene expression in S. cerevisiae crosstalk is reduced compared to an endogenous transcription factor.
Cleavage by inducible TEV protease
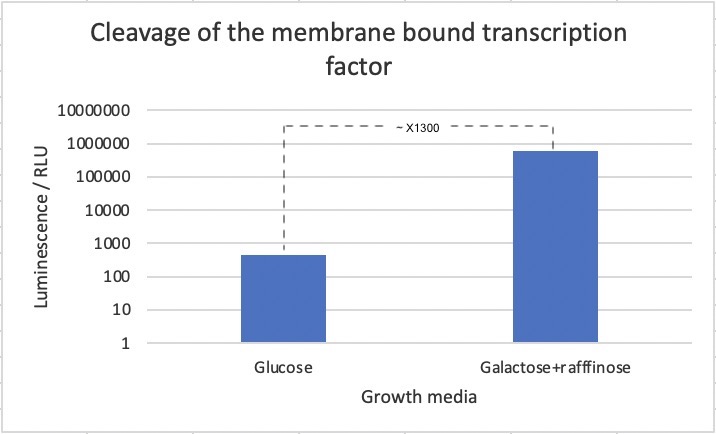
In order to test whether a non-membrane bound TEV protease is able to initiate the transcription of a reporter gene by cleaving the protein product of the biobrick, a specific S. cerevisiae strain was designed. This strain had BBa_K3617007, a TEV protease controlled by an inducible promoter; scGAL1, and a NanoLuc luciferase reporter under control of the (6x)lexAop-eno1 promoter, which as mentioned, is activated by lexA-VP16. The NanoLuc luciferase is a modified version of luciferase which luminesces 100 times brighter than firefly or Renilla luciferase.
The strain was then incubated with either glucose or galactose + raffinose media, which leads to repression or activation of TEV protease expression respectively. Following this, a luciferase assay was performed for measuring of luminescence after application of an industrial extraction reagent called YeastBuster to the samples, which allows for fast extraction of active proteins from yeast without mechanical disruption and enzymatic lysis, mixed with NanoLuc substrate which causes the luciferace to produce luminescence.
We observed a 700-fold increase in luminescence intensity after induction of the TEV protease. This strongly suggest that the part works as intended. We must mention though, that we cannot exclude that part of the difference in luciferase concentration stems from other effects of changing the media with the current experimental design. We should have used a strain with only the reporter and only inducible TEV protease and reporter as additional controls to account for other factors of changing the media. That BBa_K3617007 truly function by being cleaved by the TEV protease could be further confirmed with a westernblot of after incubation of the strain the different media. Next steps would be to investigate whether BBa_K3617007 can also be cleaved by a split-TEV protease.
A 700-fold increase in luminescence intensity was observed following expression of the TEV protease. This result infers that the biobrick is working as intended.
| None |