Part:BBa_K3349008
Pectin Lyase PelC
The PelC Pectin Lyase is originally from Paenabacillus amylolyticus. This particular pectin lyase is more specific to methylated pectin or homogalacturonan substrates rather than polygalacturonic acid [1]. This protein also works with both PelB (BBa_K3349001) and Pnl (BBa_K3349001) in this organism to effectively break down pectin. PelC has an optimal activity at 55°C and a pH of 10. The Km in terms of 80% methylated substrates was determined to be 0.41 +/-0.06mg/ml. Calcium is a cofactor for this protein and is therefore necessary within the buffer conditions [1].
For purification purposes, this gene has a C-terminal hexa-histidine tag. For more information on our design and use please see our webpage https://2020.igem.org/Team:Lethbridge_HS.
In the beginning of our project we decided to do homology modelling of our pectin lyase enzymes in order to inform our thermostability modelling.

Figure 1: Homology modelling of PelC from P. amylolyticus /i>. The protein structure was done using iTASSER [2] and SWISS-Model [3]. The two structures were then aligned and compared. A more in depth analysis can be found on our webpage.
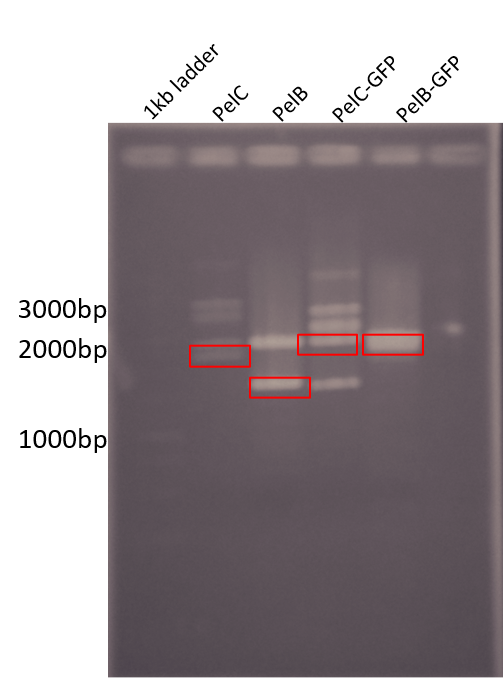
Figure 2: Restriction digest of PelB and PelC DNA from the pUC57 plasmid using EcoRI and PstI enzymes run on a 1% agarose gel. The red boxes show the band corresponding to that of our DNA constructs.
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25INCOMPATIBLE WITH RFC[25]Illegal AgeI site found at 901
- 1000INCOMPATIBLE WITH RFC[1000]Illegal BsaI.rc site found at 287
| None |
