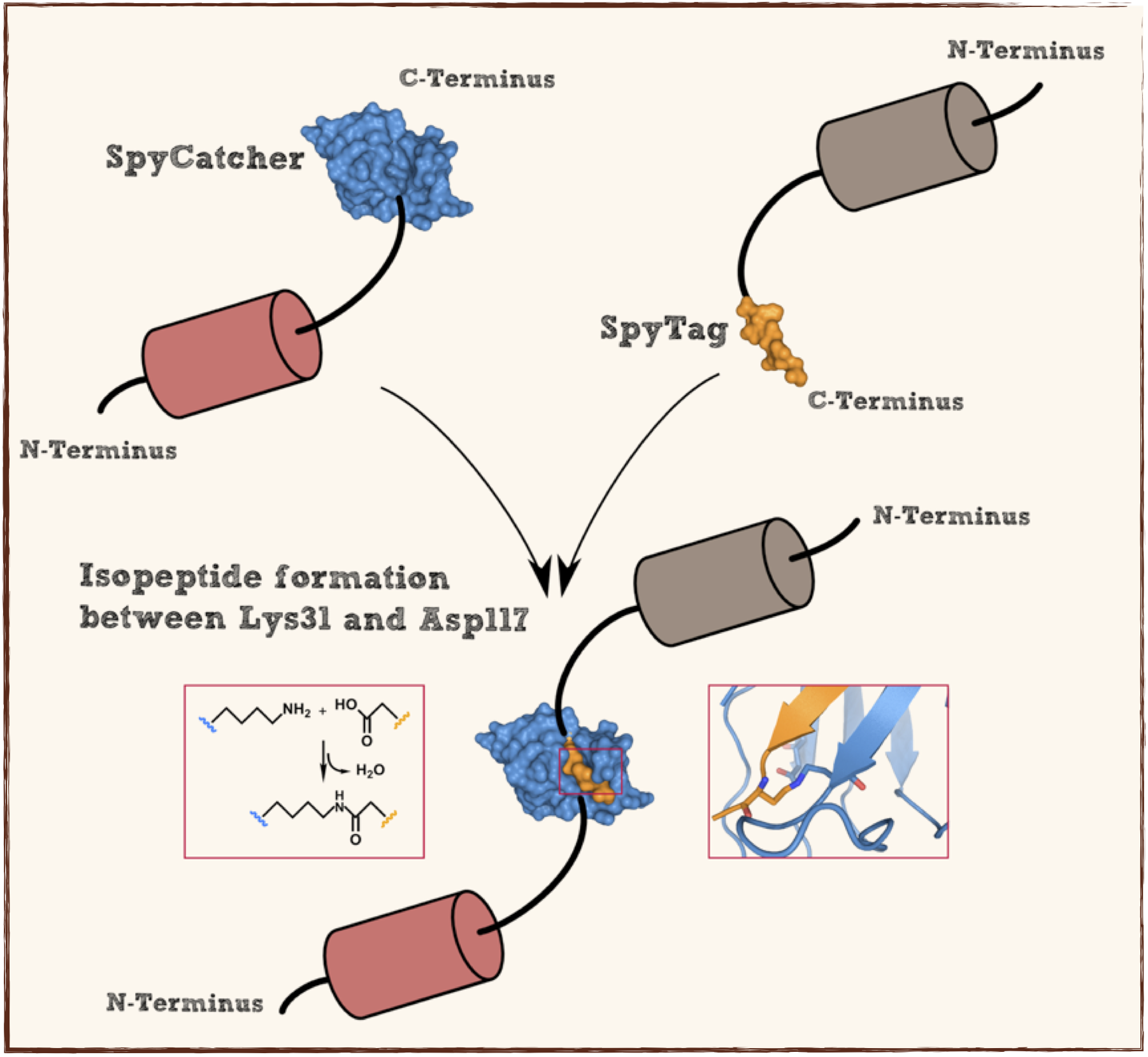
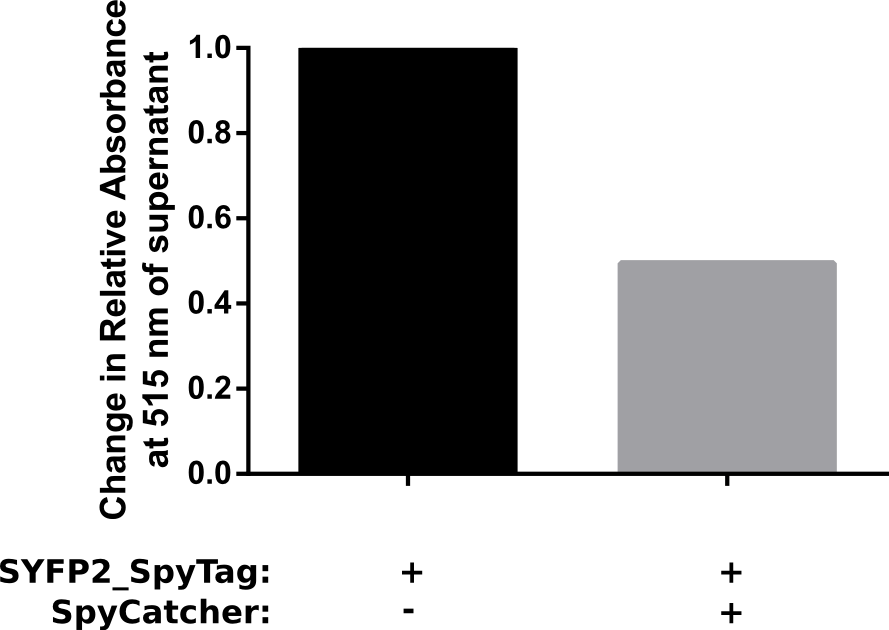Part:BBa_K1159201
Splitted and engineered C-terminal FbaB for isopeptide bound formation (SpyTag) in RFC[25]
This part codes for a oligopeptide that is recognized and bound by a covalent isopeptide bound to a protein, called SpyCatcher. That catcher protein for this oligopeptdie can be found under BBa_K1159200. This part is flanked by RFC[25] pre- and suffix for further protein fusions.
This part codes for a protein that recognizes and forms a covalent isopeptide bound to a oligopeptide. That oligopeptide can be found under BBa_K1159200. In contrast to the original SpyCatcher this part only contains the essential part for forming isopeptide bounds and does not contain a N-terminal His-tag and TEV cleavage site. Furthermore two amino acids were exchanged to improve the function, Ile34 was replaced by Glu34 which enhances the reaction rate, also the exposed hydrophobic redidue Met69 was replaced by Tyr69. This part is flanked by RFC[25] pre- and suffix for further protein fusions.
Experimental Evidence
Proper funtion of the construct was verified in a pulldown assay using 10 μM of SpyCatcher with N-terminal HisTag which was incubated with 5 μM of C-terminal spytagged SYFP2 for 1 h in PBS. Afterwards HisTag beads were added to bind the fusion product, resulting in a decrease of spytagged SYFP2 in the supernatant. The results of this assay are shown in Figure 2. The results show that after one hour at room temeperature, a two fold change in relative absorbance was observed. This verifies that the construct is working as intended.
Furthermore the two interaction partners were mixed, incubated and analyzed via SDS Page (reducing conditions) and coomassie staining.
Part Characterisation by Oxford iGEM 2019
Spy & Go Purification
Summary
We have used this part as an affinity tag to purify our proteins. To assess the quality of purification, we used mClover3 fluorescent protein fused with SpyTag and 6xHis tag and we utilised densitometric analysis on our SDS-PAGE gels.
Documentation
We adapted BBa_K1159201 as an affinity tag for our proteins. In our construct, we used the SpyTag as a secondary affinity tag. The purification method was developed by Anuar et al., 2019. It uses an immobilized protein, SpyDock which can bind SpyTag reversibly. SpyDock captures fusion tags and allows elution at high (2.5M) imidazole concentration. To assess the capabilities for purification, we used densitometric analysis to determine how pure our protein product is and how the purification relates to the widely-used 6xHis Ni-NTA purification method. We used a construct that consisted of mClover3 with SpyTag and 6xHis tag.
Methods
The samples were expressed in the same conditions (BL21 (DE3) - RIPL E. coli expression strain, pET28a expression vector with T7 expression system, induced with IPTG (420mM final concentration) at OD600 = 0.6 and at 21C. The cultures were spun down and resuspended in lysis buffer (Spy & Go: 40mM Tris, 25mM Phosphoric acid, pH = 7; Ni-NTA: 30mM Tris, 300mM NaCl, 10mM imidazole for IMAC purification; both had PI (Roche), PMSF (1mM), benzonase (1U/mL), and hen egg white lysozyme (1mg/mL) added) and lysed using sonication. After centrifuging the lysate at 17000g for 10min, the clear lysate was incubated with 250uL packed resin volume in both cases and incubated for 1 hour before loading onto a gravity column. The columns were washed 4 times, and the protein was eluted with Ni-NTA elution buffer (200mM imidazole, 30mM Tris, 300mM NaCl) or Spy&Go elution buffer (2.5M imidazole, 40mM Tris, 25mM Phosphoric acid, pH = 7.00). 500μL fractions were collected and run on an SDS-PAGE (fig. 5 & 6).
Results
Using BCA assay, we determined the total protein concentration of the samples. Then, the samples were loaded onto another SDS-PAGE gel in 0.5mg/ml concentration. The SDS-PAGE gel (run for 45 min, at 200V) was stained with Coomassie blue (InstantBlue™️) and then imaged using BioRad Chemidoc XRS and analysed using Image Lab (ver. 6.0.1, BioRad). In Image Lab, bands were detected and compared with the rest of the lane at background subtraction of disk size 2mm. Percentage purity is equivalent to Lane % (see fig. 7).
Spy&Go purification was able to purify the protein of interest with 60.1% (±4.4%, n=3) purity. This is lower than that of the Ni-NTA purification (74.5%, ±5.0%, n=3). Using this data and the results of BCA assay, we were able to determine that the total concentration of mClover3 from the Spy&Go purification was 1.0 mg/ml (±0.316mg/ml, n=3) from, while the Ni-NTA purification yielded1.5 mg/ml (±0.344mg/ml, n=3). From this, we were able to calculate the approximate yield. Spy&Go purification yielded a 8.2 mg/L (±2.525, n=3) of mClover3, whereas Ni-NTA purification yielded a 15.4mg/L of mClover3 (±3.438, n=3).
SpyTag Oligomerisation Characterization
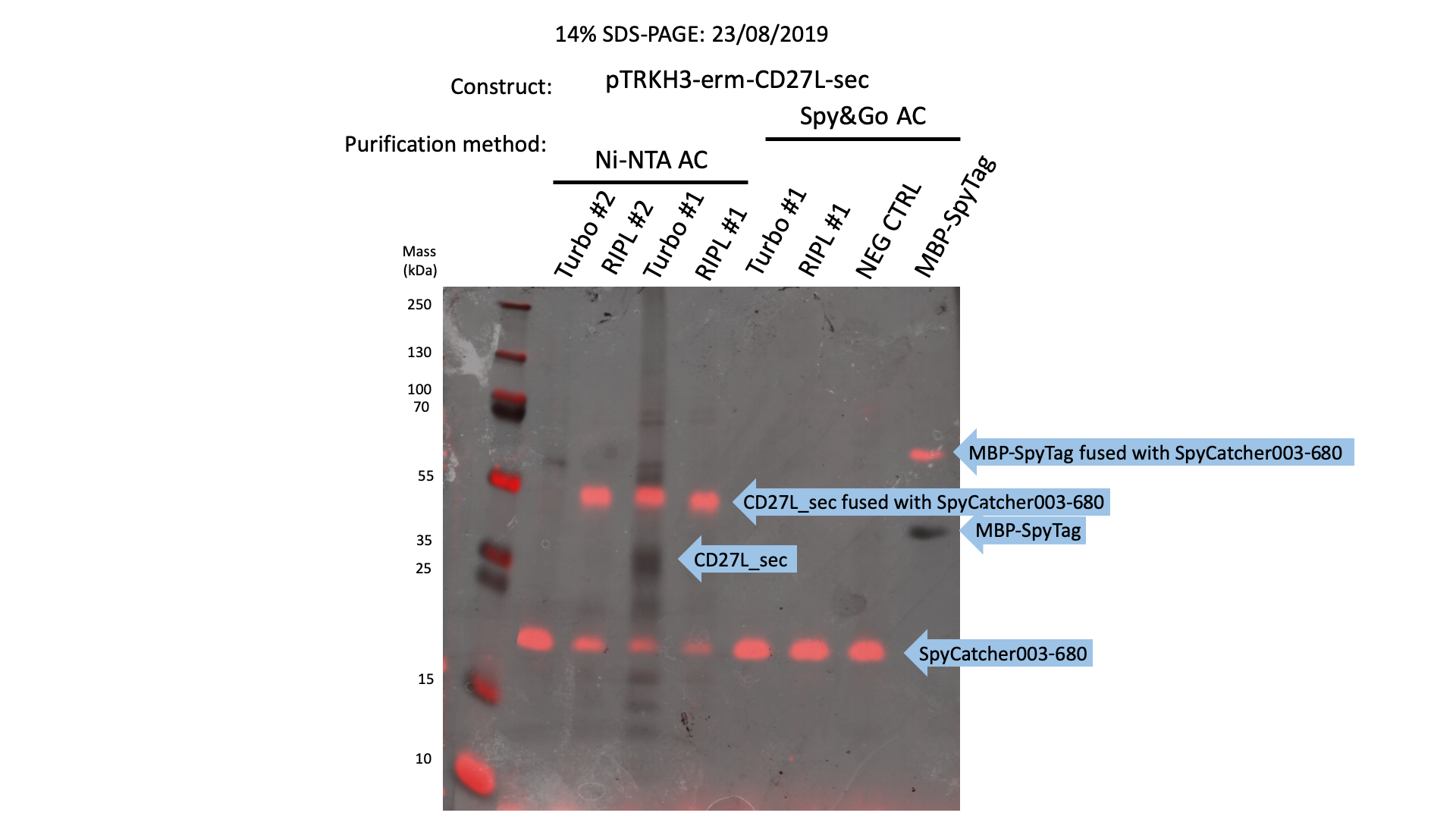
The 2019 Oxford iGEM Team used our 6His-SpyTag-CD27L construct to determine whether SpyTag can be used to fuse proteins together.
We were able to show that gel-shift assays were functional with SpyCatcher fused to a 700nm fluorophore could be used to sensitively detect protein expression (Fig. 8):
Reference
Zakeri B, Fierer JO, Celik E, Chittock EC, Schwarz-Linek U, Moy VT, Howarth M. (2012). Peptide tag forming a rapid covalent bond to a protein, through engineering a bacterial adhesin. Proc Natl Acad Sci U S A. 20;109(12)
Anuar, I. N. A. K., Banerjee, A., Keeble, A. H., Carella, A., Nikov, G. I., & Howarth, M. (2019). Spy&Go purification of SpyTag-proteins using pseudo-SpyCatcher to access an oligomerization toolbox. Nature Communications, 10(1), 1–13. https://doi.org/10.1038/s41467-019-09678-w
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000COMPATIBLE WITH RFC[1000]
| Protein data table for BioBrick BBa_ automatically created by the BioBrick-AutoAnnotator version 1.0 | ||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Nucleotide sequence in RFC 25, so ATGGCCGGC and ACCGGT were added (in italics) to the 5' and 3' ends: (underlined part encodes the protein) ATGGCCGGCGCTCATATT ... CCAACTAAGACCGGT ORF from nucleotide position -8 to 45 (excluding stop-codon) | ||||||||||||||||||||||||||||||||||||||||||||||
Amino acid sequence: (RFC 25 scars in shown in bold, other sequence features underlined; both given below)
| ||||||||||||||||||||||||||||||||||||||||||||||
Sequence features: (with their position in the amino acid sequence, see the list of supported features)
| ||||||||||||||||||||||||||||||||||||||||||||||
Amino acid composition:
| ||||||||||||||||||||||||||||||||||||||||||||||
Amino acid counting
| Biochemical parameters
| |||||||||||||||||||||||||||||||||||||||||||||
| Plot for hydrophobicity, charge, predicted secondary structure, solvent accessability, transmembrane helices and disulfid bridges | ||||||||||||||||||||||||||||||||||||||||||||||
Codon usage
| ||||||||||||||||||||||||||||||||||||||||||||||
Alignments (obtained from PredictProtein.org)
| ||||||||||||||||||||||||||||||||||||||||||||||
| Predictions (obtained from PredictProtein.org) | ||||||||||||||||||||||||||||||||||||||||||||||
Subcellular Localization (reliability in brackets)
| Gene Ontology (reliability in brackets)
| |||||||||||||||||||||||||||||||||||||||||||||
Predicted features:
| ||||||||||||||||||||||||||||||||||||||||||||||
| The BioBrick-AutoAnnotator was created by TU-Munich 2013 iGEM team. For more information please see the documentation. If you have any questions, comments or suggestions, please leave us a comment. | ||||||||||||||||||||||||||||||||||||||||||||||
| None |