Part:BBa_K2406020
T7-LacO Promoter
Introduction
T7 Promoters are promoters that work with T7 RNA Polymerase. T7 RNA Polymerase is commonly used in biotechnology, as evidenced by the large number of parts found in the iGEM registry. T7 Polymerase has high affinity for its associated promoters and was identified as a useful tool for producing high amounts of RNA back in the 1980’s [1]. T7 Polymerase is also non-natively expressed in E. coli. Our team aimed to take advantage of this fact to decrease leaky expression, as explained in the figure below. If T7 polymerase is expressed inducibly under the control of a LacI promoter, and the protein of interest is transcribed under the control of an inducible T7 promoter, you are unlikely to get high amounts of “leaky” production of your protein of interest. This is because it would require two simultaneous “leaks” at two inducible promoters, rather than just one leak. Clearly, T7 Polymerase is a useful promoter to add to the synthetic biologist’s repertoire. Therefore, we were surprised to see there were no inducible T7 Promoters that were well-documented and verified to function in the registry. BBa_R0184,BBa_R0185,BBa_R0186, and BBa_R0187 were all supposed to be inducible T7 promoters, yet they had no functional verification. We sought to improve these parts by developing our own T7 promoter that could be induced.
Results
We designed our T7 promoter so that it could be induced by IPTG. Therefore, we simply ligated the Lac Operator sequence to the popular BBa_I712074 constitutive T7 promoter. Sequencing results (accessible by download and linked on this page) showed that we had successfully attached the T7-LacO promoter to our recombinase protein expression units, creating inducible expression generators (BBa_K2406080, BBa_K2406081, BBa_K2406082, BBa_K2406083, BBa_K2406084). We assayed the inducible activity of each recombinase generator. The image below shows the activity of Dre Recombinase protein. Focus on bars indicating Rox-Rox constructs (BBa_K2406051 part has more information on how this assay works), which were tested with and without IPTG. Essentially, RFP would be expressed and give off fluorescence when Dre recombinase expression was induced. Clearly, there is a significant difference between induced and non-induced control, demonstrating that our T7-LacO promoter works as would be desired.
Discussion
There was some leakiness observed by the T7-LacO promoter. Comparing un-induced Rox-Rox construct to other controls that did not give RFP output with or without Dre recombinase, there is clearly significantly more RFP fluorescence in the un-induced Rox-Rox construct. This indicates some Dre recombinase being produced despite no induction occurring. As explained in the introduction, we attempted to decrease leakiness by inducing T7 LacO expression as well. Leakiness could have perhaps been further reduced by using a straing carrying pLysS plasmid, which would express lysosyme to degrade any T7 polymerase produced through leakiness. Furthermore, other teams could attempt to alter the sequence of our promoter, perhaps by decreasing affinity for T7 RNA polymerase, in an attempt to decrease the intrinsic leakiness of this promoter. However, we have clearly produced an inducible T7 promoter that functions, therefore improving parts BBa_R0184, BBa_R0185, BBa_R0186, and BBa_R0187, all of which had slightly different sequences and no evidence of functioning.
References
Milligan, J.F., Groebe, D.R., Witherell, G.W., and Uhlenbeck, O.C. 1987. “Oligoribonucleotide synthesis using T7 RNA polymerase and synthetic DNA templates”. Nucleic Acids Research 15 (21): 8783-8798
Sequences
Find sequences of all promoter-regulated generators and measurement constructs here: Media:File:Sequencing Results Edinburgh UG.zip
Team Tec-Monterrey characterization
While we were working in the expression of our chimeric proteins we realized that the promoter has an important leakage.
We first noticed visually by noticeable fluorescence on non-induced bacteria.
Figure 1. 200µM IPTG induced sfGFP transformed bacteria (note that it flouresces less than the non-induced one).

Figure 2. non-induced E.coli.
We confirmed the leakage with a fluorescence measurement, where bacteria were growth at 30ºC for 3 hrs. Then 100µl of the grown bacteria were induced with IPTG at 0, 40, 200 and 400 mMm microtiter florescent measures were taken every half an hour for a period of 3 hours. Absorption and emission values were 395/509 nm.
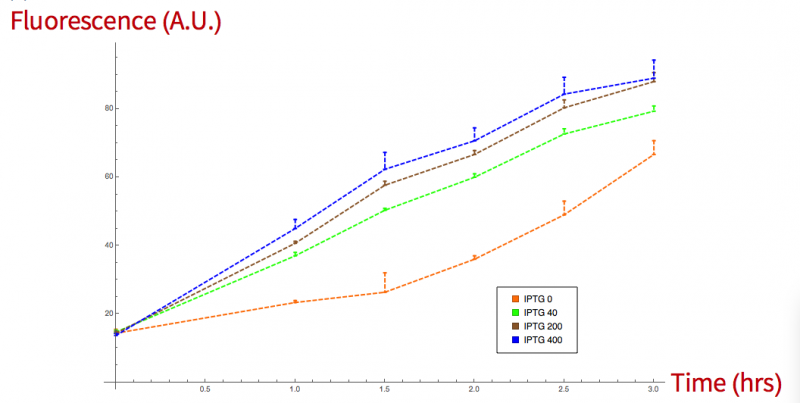
Figure 3. sfGFP grpah, fluorescence vs time induced at different concentrations.

Figure 4. sial+sfGFP grpah, fluorescence vs time induced at different concentrations.
NOTE: We suggest that the lower fluorescence values in the induction of the sfGFP+sialidase than the sfGFP itself are due to the different sizes of the chimeric proteins that could difficult its secretion.
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000COMPATIBLE WITH RFC[1000]
| None |


