Part:BBa_I712019
Firefly luciferase - luciferase from Photinus pyralis
Firefly Luciferase is luciferase from Photinus pyralis. By oxidation of substrate luciferin light is produced. Luciferase thus acts as a reporter protein when connected to other proteins or promoters.
Characterization by Team Munich 2019
In iGEM 2019, we the iGEM Munich team, characterised the Firefly Luciferase.
Aim:
Determination of equilibrium parameters of the wild-type Firefly Luciferase (Part:BBa_I712019) in vivo in mammalian cells using the cell-permeable substrate, D-Luciferin.
Design:
As a first step, we ordered the sequence for the wild-type Firefly Luciferase (Part:BBa_I712019) and ligated it into a backbone under the control of a CAG promoter optimised for HEK293T cells (BBa_K3113200) using the Gibson Assembly technique. After sequencing confirmation, we transfected HEK293T cells with the plasmid containing the sequence for Firefly Luciferase (pCAG_FLuc). Finally, we performed a luciferase assay with different D-Luciferin concentrations and measured the luminescence with a plate reader.
Materials and Measurement:
- pCAG_FLuc
- HEK293T cells
- D-Luciferin (100 mM stock solution)
- GloMax® Discover Microplate Reader (Promega)
HEK293T cells were transfected with pCAG_FLuc using our standardized protocol for transfection using Lipofectamine P3000. The cells were then grown for 48 hours at 37 °C and 5 % CO2. A dilution series of D-Luciferin was prepared ranging from 0 μM up to 8 mM, which was then added 1:1 to the transfected cells, reaching final concentrations from 0 μM up to 4 mM. As negative control we included samples where no substrate was added. For the luciferase assay we used the GloMax® Discover Microplate Reader to measure the luminesence of our transfected cells. The assay was carried out at 37°C and no additional ATP or Mg2+ was added. For each substrate concentration we included four biological replicates, which were then averaged. D-Luciferin has a half-life of over 24 hours, allowing long-term measurements. Therefore, we measured our samples every five minutes over a period of approximately 8 hours, gaining more data for technical replicates.
Analysis:
All the analyses were performed using Graphpad Prism (San Diego, CA).
Results:
Time-resolved data of biological replicates for the luciferase assay are summarized in Figure 1. We observed a concentration dependency of Firefly Luciferase in regard to D-Luciferin.

We determined the equilibrium binding constant, Kd (D-Luciferin), by plotting the averaged equilibrium values against the used D-Luciferin concentrations. The calculated equilibrium binding constant Kd (D-Luciferin) equals 0.9619 mM, representing the D-Luciferin concentration needed to achieve half-maximum binding at equilibrium. The solid line represents the fit used for the determination of the equilibrium binding constant.
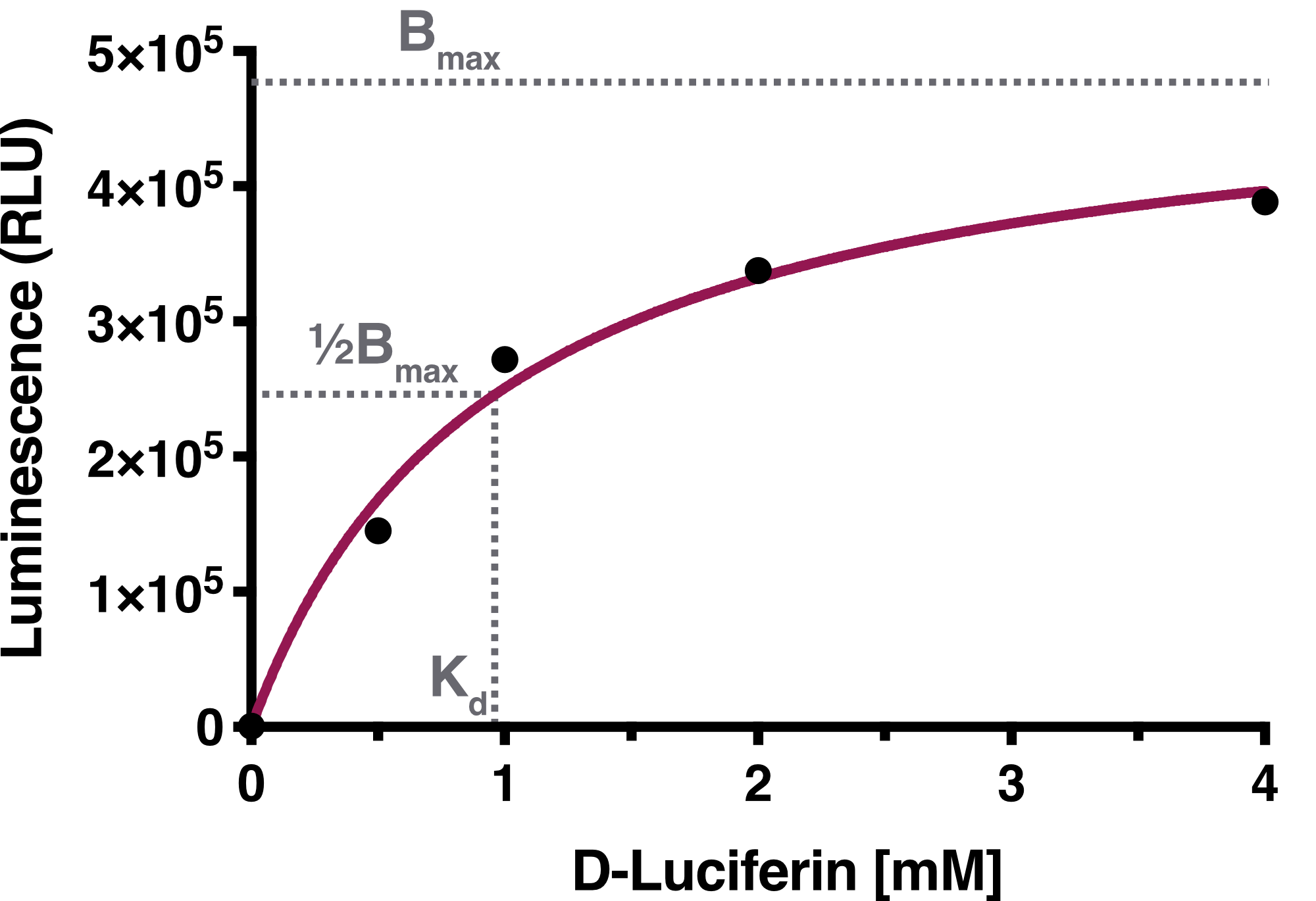
Discussion:
By following the luminescence signal over a longer time period and including four biological replicates, we obtained consistent data. We aimed to standardize this assay by establishig a standart curve with purified Firefly Luciferase from E.coli, but unfortunately we weren't able to successfully purify FLuc overexpressed from E.coli cells. However, we standardized our assay by seeding the same amount of cells into each well and transfecting them with the same concentration of DNA and included four biological replicates for each condition tested.
Modification
In iGEM 2016, our team, SYSU-MEDICINE, had constructed a new composite part BBa_K1993006 in order to improve the positioning function of this previous part. Our new composite part contains gene Firefly Luciferase, dTomato and hFTH, which help users observe biological processes in vivo and in vitro.
For more information, please see our Part page: https://parts.igem.org/Part:BBa_K1993006
Improvement of the function
This part was improved by NUDT_CHINA in iGEM 2017 to optimize its function in a dual-luciferase miRNA target expression system. Two restriction sites of BsmBI were added to the 3’ end of firefly luciferase, through which, the 3’ untranslated region of any gene can be inserted using Golden-Gate Assembly, with no worry about the multiple illegal sites contained in it. For more information, please visit our part page: https://parts.igem.org/Part:BBa_K2440008.
Usage and Biology
This part contains a Eukaryotic ribosome binding site and non-standard prefix and suffix. See the sequence first.
We used the part BBa_I712019 to construct the part with split-luciferase. Our sequencing result suggested that this part is completely working. --------Tianjin IGEM TEAM
In iGEM 2016, our team, SYSU-MEDICINE, had constructed a new composite part BBa_K1993006 in order to improve the positioning function of this previous part. Our new composite part contains gene Firefly Luciferase, dTomato and hFTH, which help users observe biological processes in vivo and in vitro.
For more information, please see our Part page: https://parts.igem.org/Part:BBa_K1993006
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000INCOMPATIBLE WITH RFC[1000]Illegal SapI.rc site found at 808
//function/reporter/light
| None |

 1 Registry Star
1 Registry Star