Part:BBa_K1104200
OxyR
Introduction
OxyR is Escherichia coli transcription factor that senses H2O2 to activate transcription, and is activated through the formation of an intramolecular disulfide bond. The OxyR gene, a positive regulatory gene for the oxidative response, then controls proteins which are induced by treatment with H2O2.

Literature Study of OxyR
OxyR, a positive regulator of hydrogen peroxide-inducible genes in E. coli. The OxyR transcription factor regulates the H2O2 response of E. coli.
Oxidative stress, through the production of Reactive Oxygen Species (ROS) , is a natural consequence of aerobic metabolism. Escherichia coli has several major regulators activated during oxidative stress, including OxyR, SoxRS, and RpoS. OxyR undergo conformation changes when oxidized in the presence of hydrogen peroxide, then positively controls the expression of cognate genes.
- Reference: Chiang, S. M., Schellhorn, H. E., & Catalases and Hydrogen Peroxide Metabolism. (September 15, 2012) . Regulators of oxidative stress response genes in Escherichia coli and their functional conservation in bacteria. Archives of Biochemistry and Biophysics, 525, 2, 161-169.
H2O2 oxidizes OxyR Cys199 to an R-SOH that reacts with Cys208 in an intramolecular disulphide bond.
- Reference: D'Autréaux, B., & Toledano, M. B. (October 01, 2007). ROS as signalling molecules: mechanisms that generate specificity in ROS homeostasis. Nature Reviews Molecular Cell Biology, 8, 10, 813-824.
Usage and Biology
After Nosema ceranae enter bees' midgut, the natural immune reaction will lead to ROS released by epithelia of bees' midgut. In our project: Bee. coli, we use OxyR-induced promoter as the sensor of Bee. coli to sense Nosema ceranae. We select several promoters that can be induced by OxyR(Part:BBa_K1104200).
There are promoters that can be activated by OxyR: TrxCp(Part:BBa_K1104201), HemHp (Part:BBa_K1104202) , ahpC (Part:BBa_K362001) , sufA (Part:BBa_K362005) , and dps (Part:BBa_K362002) , yiaA (Part:BBa_K362006) .
The OxyR-induced promoters:
- TrxCp (Part:BBa_K1104201) and HemHp (Part:BBa_K1104202) are parts created by our team [http://2013.igem.org/Team:NYMU-Taipei/Modeling/MainParts NYMU_Taipei]. (Come to the [http://2013.igem.org/Team:NYMU-Taipei/Experiment/Wet_Lab Wet Lab] page to see our part list.)
- sufA (Part:BBa_K362005) , designed by [http://2010.igem.org/Team:KIT-Kyoto/Parts 2010 KIT-Tokyo team], is successfully characterized again by us.
- ahpC (Part:BBa_K362001) is improved its complex composition and Assembly Compatibility by us. We designed ahpC (Part:BBa_K362001) improvement into 6 versions:
| AhpCp1000 (Part:BBa_K1104203) | The PstI cutting site is mutated. |
| AhpCp2D1 (Part:BBa_K1104204) | After mutating the PstI cutting site, the truncated coding sequence from the DsbG promoter sequence is removed. |
| AhpCp2 (Part:BBa_K1104205) | Only one promoter(AhpCp2) and its TFBS. |
| AhpCpD1 (Part:BBa_K1104206) | Bidirectional promoter: AhpCp1 and DsbGp(reverse promoter), and their shared TFBS. |
| AhpCp1 (Part:BBa_K1104207) | Only one promoter(AhpCp1) and its TFBS. |
In order to enhance the strength and sensitivity of ROS-sensing promoters, we make E. coli continuously produce inactivated OxyR transcription factors, so that ROS can affect promoters more easily.
We designed a device that is composed of a sensor (OxyR-induced promoter, including TrxCp, HemHp, sufA, AhpCp1000, AhpCp2D1, AhpCp2, AhpCpD1, AhpCp1, DsbGp) plus reporter (Part:BBa_E0840). Device1 in order to enhance the effect of ROS on E. coli is added ahead: a constitutive promoter(Part:BBa_J23102), and OxyR(Part:BBa_K1104200).
Contribution: Improvement and Characterization
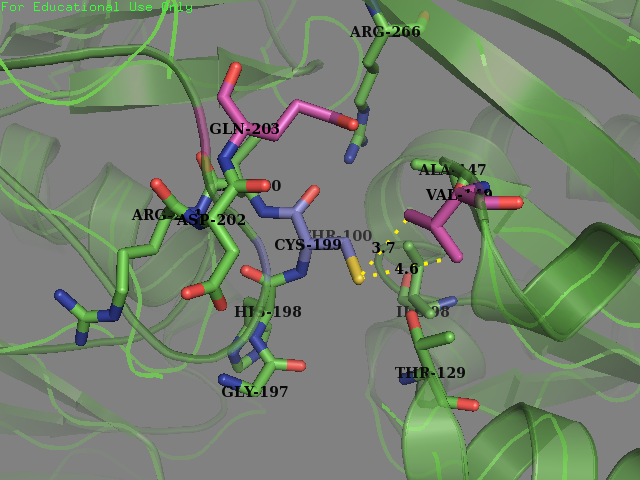
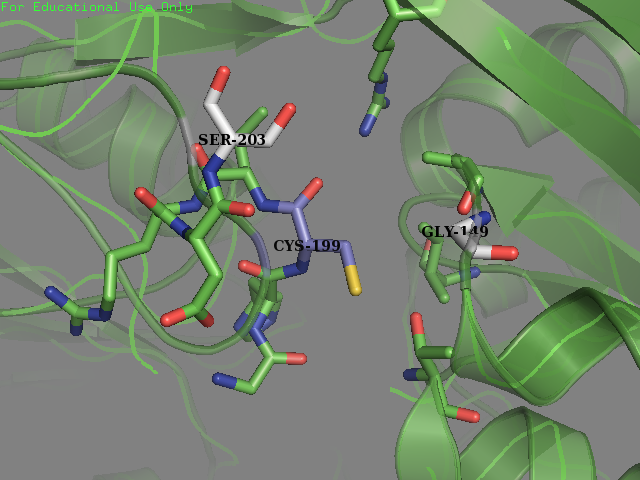
Team SUIS_Shanghai 2019 attempted to improve the sensitivity of this OxyR to reactive oxygen species (ROS) by increasing the accessible surface area of the reactive cysteine (cysteine-199) within the protein. This cysteine, once oxidized will form a intermolecular disulfide bond with Cys-208, resulting in a conformational change which can activate certain ROS sensitive promoters and therefore positively expressing genes in times of oxidative stress. Our approach was to use a large set of statistical data to identify certain structural features within the environment of reactive cysteines in a wide variety of protein structures sourced from the Protein Data Bank (PDB). We compared the structural features of these oxidized cysteine environments (the cysteines will form a Sulfenic acid upon oxidation – a post translational modification which is known to be a switch in some proteins) with unmodified cysteine residues within the same proteins. The information we obtained from this large study (of over 300 different proteins) was then used to inform decisions into editing the OxyR gene sequence to mutate the amino acid sequence around the Cys-199. We hoped to make this transcription factor more sensitive to lower concentrations of H2O2 and potentially create a new device which could be used to be a very sensitive indicator of early immune response and infection.
To obtain a comprehensive account of the data used to make the decisions to edit the OxyR nucleotide sequence, please visit our Model page. Our analysis led us to change two amino acids in the OxyR protein surrounding the Cys-199 and create a new part BBa_K3031018.
We mutated amino acids GLN-203 and Val-147 in the original protein.
We sent our new sequence to Genscript China to be sequenced, along with the original OxyR sequence. The circuit we used to measure any improvement in our cells is found below:
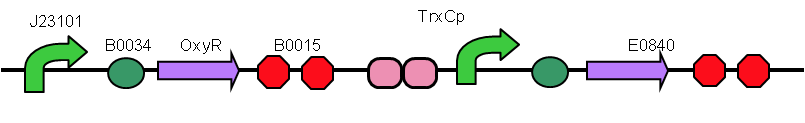
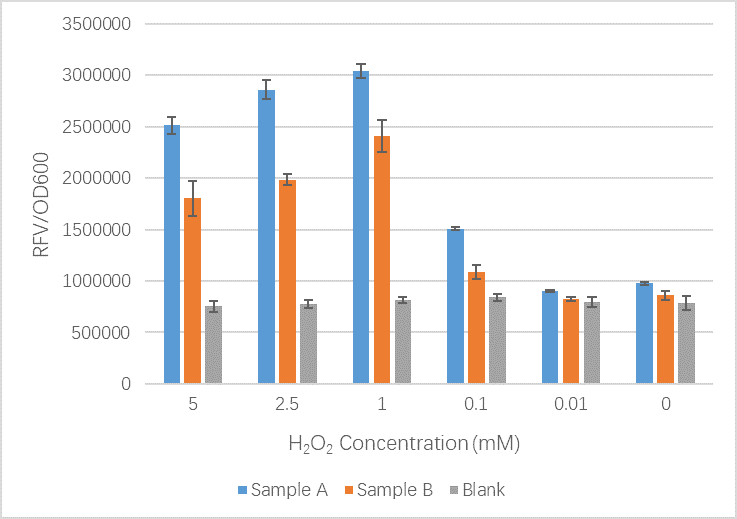
The OxyR gene is expressed by a constitutive promoter while the inducible promoter TrxC will only express downstream genes (GFP in this case) when it is activated by oxidized form of OxyR, by virture of binding to sites (maked in pink here). When H2O2 levels are high enough the OxyR is activated and GFP is produced. Both constructs were sequenced onto a expression vector pET301(+) and transformed the plasmid into Bl21(DE3) cells. To induce the TrxC promoter we divided four 50ml test tubes of OxyR and OxyR_Mutated into twelve 15ml test tubes, each contained 5ml culture broth and 5ml LB with OD600 0.4. H2O2 was added to 10 of the tubes in the following concentrations:

Where:
- Sample A = E.coli BL21(DE3) cells containing expression plasmid pET30a(+) containing: Composite part with original gene sequence of OxyR (this part) under influence of constitutive promoter and GFP under the influence of inducible promoter TrxCp
- Sample B = E.coli BL21(DE3) cells containing expression plasmid pET30a(+) containing: Composite part with mutated gene sequence of OxyR (BBa_K3031018) under influence of constitutive promoter and GFP under the influence of inducible promoter TrxCp
- Blank = E.coli BL21(DE3) cells containing expression plasmid pET30a(+) containing no engineered plasmid.
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000COMPATIBLE WITH RFC[1000]
Related Parts
| ahpC(Part:BBa_K362001) | |||||
| AhpCp1000 (Part:BBa_K1104203) | AhpCp2D1 (Part:BBa_K1104204) | AhpCp2 (Part:BBa_K1104205) | AhpCpD1 (Part:BBa_K1104206) | AhpCp1 (Part:BBa_K1104207) | DsbGp (Part:BBa_K1104208) |
| AhpCp1000+E0840 (Part:BBa_K1104243) | AhpCp2D1+E0840 (Part:BBa_K1104244) | AhpCp2+E0840 (Part:BBa_K1104245) | AhpCpD1+E0840 (Part:BBa_K1104246) | AhpCp1+E0840 (Part:BBa_K1104247) | -- |
Sequence and Features
| None |


