Part:BBa_K3196011
AOX1-Kozak-FLO10-SLAC-His tag-AOX1 Terminator
Characterization
This is a composite part that used to degraded lignin. SLAC is a multicopper oxidase isolated from S. coelicolor , capable of catalyzing one-electron oxidation of a wide range of substrates to generate radicals while concomitantly reducing molecular oxygen to water[1].
FLO10 is signal peptide suitable for expression in pichia pastoris Lectin-like protein with similarity to Flo1 p,thought to be expressed andinvolved in flocculation.[2]
P. pastoris is usually the preferred host for the production of industrial enzymes.
DNA Gel Electrophoretic
To confirm the function of this part, first we confirm that the gene is transferred to P. pastoris GS115 successfully.
1.DNA extraction of the E.coli plasmid and verification of the right fragment.
2.Prepare the competent cells of P. pastoris GS115.
3.Electro transformation.
4.Yeast genome extraction and PCR verification.
As the picture shows, we have constructed the engineering bacteria successfully.
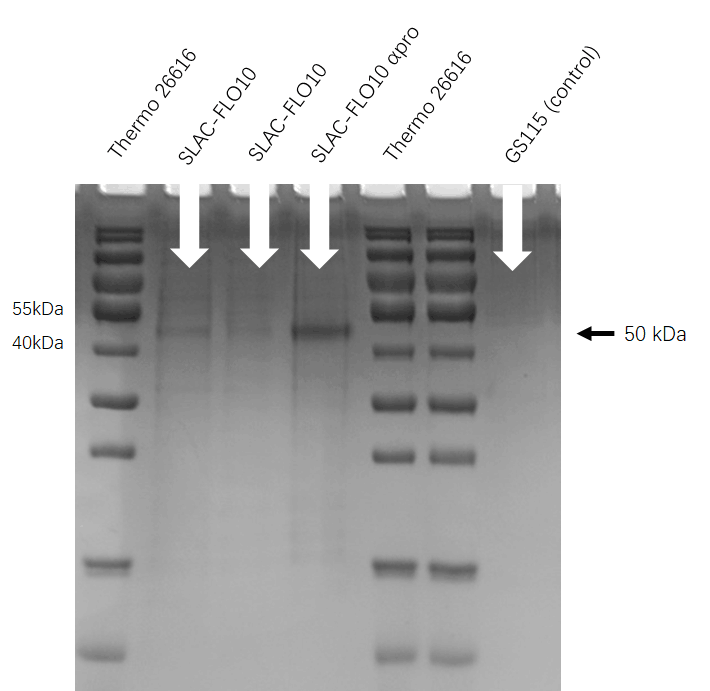
SDS-PAGE
Second, we cultured the engineering P. pastoris GS115(FLO10-SLAC)in the buffered glycerol-complex medium (BMGY) and induced it in buffered minimal methanol medium (BMM).
Enzyme Activity
Laccase activity was determined at room temperature (22–25 °C) using ABTS. Oxidation of ABTS (1 mM) was measured at 420 nm (ε = 36,000 M−1 cm−1) in 20 mM acetate buffer (pH 4.0).
By using this formula: 〖activity=(A2−A1)〗∕t∗11244
We obtain the follow figure that represent the enzyme activity changes with time.
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21INCOMPATIBLE WITH RFC[21]Illegal BamHI site found at 937
Illegal BamHI site found at 948 - 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000INCOMPATIBLE WITH RFC[1000]Illegal BsaI.rc site found at 1387
| None |