Part:BBa_K3078005
LL-37
LL37 protein coding region. We added a stop codon to BBa_K1162006.
1. Usage and Biology
LL-37 is an antimicrobial peptide (AMP) from the cathelicidin family of antimicrobial peptides, isolated from the Human (Homo sapiens). It is a 37-residue helical peptide found throughout the human body, exhibiting a broad spectrum of antimicrobial activity as well as a variety of immunomodulating effects (Dürr, Ulrich H.N., et. al 2006). It has the distinction of being the first and only cathelicidin member discovered from humans, and is extremely well studied and characterized for this reason. The peptide is produced via epithelial cells as well as leukocytes in places such as the skin, gastrointestinal tract, and the respiratory tract. This AMP is is cleaved from a larger protein, hCAP-18, in a series of modifications to yield its active form (Sorensen, O.E, et. al 2001).
2. Characterization
2.1 Validation of pVE-β-1,3-glucanase construction
We characterised β-1,3-glucanase by cloning it into pVE vector. Moreover, an signal peptide were added.
To verify the construction of pVE-β-1,3-glucanase(pVE-β-GA) which we generated, the digestion by SalI/EcoRV was performed by a standard protocol following agarose gel electrophoresis (Figure 1).
Figure 1. Digestion and agarose gel electrophoresis of pVE-β-GA.
2.2 Expression of pVE-β-1,3-glucanase
To assess the expression of our β-1,3-glucanase, Congo Red experiment was used.
Congo Red has a strong chromogenic reaction with β-1, 3-glucan, and when β-1, 3-glucan is decomposed into reducing monosaccharides by β-1, 3-glucanase, the hydrolyzed region forms a pale yellow transparent hydrolytic circle. The results of activity detection of Congo Red hydrolytic ring (Figure 2) showed that compared to the control, there was a larger size of transparent hydrolytic circle caused by β-1, 3-glucanase expressed in E. coli with pVE-β-GA.
Figure 2. Expression of pVE-β-GA. Add 10 mg/ml Congo Red solution to LB medium containing β-1,3-glucan substrate (0.1 g/100 mL) at a ratio of 1:100. 100 μl supernatant obtained by centrifugation after ultrasonic crushing of E. coli with pVE-β-GA is added to the Oxford cup, and the pVE empty vector bacteria supernatant is used as the control. Stand at 37 ℃ for 24 hours.
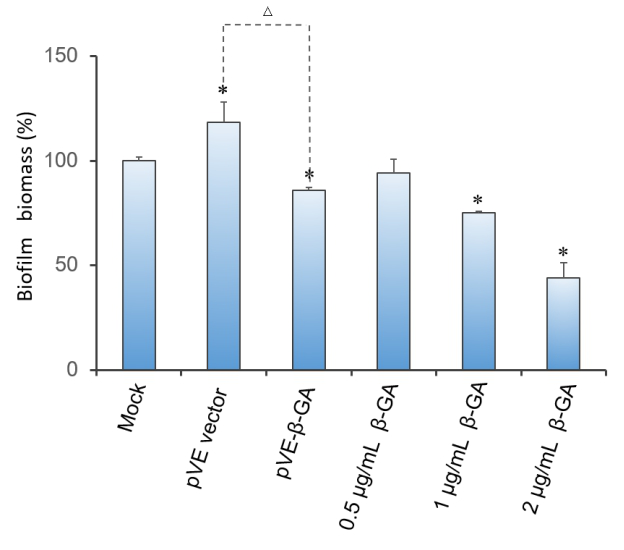
2.3 Effect of β-1,3-glucanase on biofilm in 96-well microplate
Crystal violet(CV) reduction method, which is commonly used for quantitative analysis of biofilm, was used to evaluate the antibiofilm activity of β-1,3-glucanase. The result indicates that β-1,3-glucanase has disruption effect on mature biofilm with concentration-dependent manner. Under the condition of using bacteria with pVE empty vector to exclude the influence of bacterial substances on the staining results, as Figure 3 shows, β-1,3-glucanase produced by our engineered bacteria has the effect of degrading biofilm. The supernatant of E. coli with pVE-β-GA diluted by one time is estimated to reach the effect of 0.5 μg/mL~2 μg/mL samples of standard β-1,3-glucanase.
Figure 3. Effect of β-1,3-glucanase on biofilm in 96-well microplate. Biofilm formed in RPMI 1640 medium for 48 h. Mature biofilm was treated with RPMI 1640 medium, supernatant of bacteria with pVE empty vector or pVE-β-GA and standard β-1,3-glucanase in different concentrations (0.5, 1 and 2 μg/mL) for another 24 h. Values obtained are given as the percentage of biofilm. The experiment was performed three times in triplicate. *, P < 0.05 from mock control using Student’s t test. △, P < 0.05.
3. Conclusion
Our engineered bacteria successfully characterized β-1,3-glucanase,moreover, it was effective in performing the function of degrading C. albicans biofilm.
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25INCOMPATIBLE WITH RFC[25]Illegal NgoMIV site found at 274
Illegal NgoMIV site found at 483
Illegal NgoMIV site found at 622 - 1000COMPATIBLE WITH RFC[1000]
| None |