Part:BBa_I0500
Inducible pBad/araC promoter
pBad is an E. coli promoter that is tightly controlled by:
- inducer: L-[http://openwetware.org/wiki/Arabinose arabinose].
- repressor: AraC acts as the repressor
one [http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?db=pubmed&cmd=Retrieve&dopt=Abstract&list_uids=7768852 study] concluded that [http://openwetware.org/wiki/Arabinose arabinose] can change the conformation of araC and prevent it from successfully binding to and repressing pBad.
Usage and Biology
- When grown with 0.2% arabinose, promoter is weak-medium. [jb, 5/24/04] Part may not be compatible with MC4100 as cell line is araD 139.
- MC4100 is not a good chassis for operating BBa_I0500 (pBad promoter). The feed-forward regulation of the endogenous promoter controlling expression of the arabinose transporter prevents linear induction with increasing arabinose concentration. ((Engineered strain from Keasling's lab, used by jrk for operation of the screening plasmid.))
>Internal Priming Screening Characterization of BBa_I0500: Has 1 possible internal priming site between this BioBrick part and the VF2 primer.
The 2018 Hawaii iGEM team evaluated the 40 most frequently used BioBricks and ran them through an internal priming screening process that we developed using the BLAST program tool. Out of the 40 BioBricks we evaluated, 10 of them showed possible internal priming of either the VF2 or VR primers and sometime even both. The data set has a range of sequence lengths from as small as 12 bases to as large as 1,210 bases. We experienced the issue of possible internal priming during the sequence verification process of our own BBa_K2574001 BioBrick and in the cloning process to express the part as a fusion protein. BBa_K2574001 is a composite part containing a VLP forming Gag protein sequence attached to a frequently used RFP part (BBa_E1010). We conducted a PCR amplification of the Gag-RFP insert using the VF2 and VR primers on the ligation product (pSB1C3 ligated to the Gag + RFP). This amplicon would serve as template for another PCR where we would add the NcoI and BamHI restriction enzyme sites through new primers for ligation into pET14b and subsequent induced expression. Despite gel confirming a rather large, approximately 2.1 kb insert band, our sequencing results with the VR primer and BamHI RFP reverse primer gave mixed results. Both should have displayed the end of the RFP, but the VR primer revealed the end of the Gag. Analysis of the VR primer on the Gag-RFP sequence revealed several sites where the VR primer could have annealed with ~9 - 12 bp of complementarity. Internal priming of forward and reverse primers can be detrimental to an iGEM project because you can never be sure if the desired construct was correctly inserted into the BioBrick plasmid without a successful sequence verification.
For the BioBrick part BBa_I0500, the location of the internal priming site is on the 1151-1158 base number of the BioBrick and on the 5-12 base number of the VF2 primer.
[http://2010.igem.org/Team:Slovenia Team Slovenia 2010] further characterized pBAD promoter. Check results on Experience.
[http://2016.igem.org/Team:IISc_Bangalore IISc Bangalore 2016] showed catabolite repression of expression from the promoter by glucose. Check results on Experience.
[http://2016.igem.org/Team:OUC-China OUC-China 2016] characterized BBa_I0500 of different concentrations of L-arabinose on the transcriptional level. Check results on Experience.
[http://2017.igem.org/Team:TU-Eindhoven TU-Eindhoven 2017] optimized the expression on by testing 3 different temperatures, where 37 degrees Celcius proved most successful. Addition of L-arabinose has also been optimized to compensate for the breakdown of L-arabinose by the E.coli strain BL21(DE3) for mCherry and GFP expression. Check results on Experience.
[http://2017.igem.org/Team:Glasgow Team Glasgow 2017] improved this part by splitting its constituent parts into two separate BioBricks: BBa_K2442101 encoding minimal pBAD and BBa_K2442104 encoding AraC under regulation of LacI-regulated promoter. This allows for greater control over arabinose-inducible systems. See details on Experience.
- [http://openwetware.org/wiki/Titratable_control_of_pBAD_and_lac_promoters_in_individual_E._coli_cells#pBAD_promotersOpenWetWare From an OWW article on pBAD and lac promoters]:
- Import of arabinose into cells is mediated by the araE gene. Induction of the arabinose transporter encoded by araE can be uncoupled from the endogenous PBAD promoter by deleting the chromosomal araE gene and replacing it with a plasmid-borne copy of araE under control of a constitutive promoter (1). However, this does not seem to be enough to allow for homogenous expression from PBAD promoters in a population of cells (2).
- At low concentrations of arabinose, degradation of the sugar within cells also effects the homogeneity of expression from PBAD promoters (2). Arabinose degradation is mediated by the araBAD genes. Strains lacking functional araE, araFGH (another transporter), and araBAD can be made to be responsive to arabinose for PBAD promoter induction (2). This is achieved by introduction of a mutant lacY gene. LacY A177C allows for downhill transport of arabinose, as well as maltose, palatinose, sucrose, and cellobiose (3), but does not actively transport these sugars (4). Lactose import is not affected in this mutant. So, PBAD promoters in cells lacking endogeneous arabinose importers and containing LacY A177C are linearly responsible to arabinose at the individual cell level.
- By the way, AraC is the repressor of the PBAD promoter. It is encoded on the pBAD vector series and is still present in the above-described strains.
PC and AraC are located on the complementary strand, reading right to left as written.
- At least one registry stock contains a deletion of the C at base 1194. This is after the transcriptional start but before the translation start, so it may not be significant. Parts with this mutation have been qualitatively observed to function normally.
Induction and Subsequent Inhibition of the pBAD Promoter
(Characterized by SDU-Denmark 2017)
Expression by the pBAD promoter can be regulated tightly by induction and subsequent inhibition.
The pBAD promoter holds great potential to control the expression of genes that require tight regulation, as it is capable of both an induction and repression. The [http://2015.igem.org/Team:HKUST-Rice, HKUST-Rice iGEM team] from 2015 found that the pBAD promoter exhibits an almost all-or-none behaviour upon induction with arabinose when located on a high copy vector, but allows for gradual induction when cloned into a low copy vector. Thus, it was evident that this promoter on a high copy vector would be inappropriate for tightly regulated gene expression. Based on these findings, a low copy vector was used to investigate the ability to inhibit gene expression subsequent to induction of pBAD.
Gene expression was simulated by fluorescence microscopy using a pBAD-YFP reporter system, BBa_I6058.
For this purpose, an Olympus IX83 with a photometrics prime camera was used with an exposure time for YFP at 200 ms. Transformed E. coli MG1655 cells were cultured in M9 minimal medium supplemented with 0.2% glycerol and 30 µg/mL chloramphenicol, to avoid catabolite repression from glucose residues present in LB medium. Two cultures were incubated, of which one was induced with 0.2 % arabinose from the beginning. At OD600=0.1, designated time 0, the cultures were split in two and 0.2 % glucose was added to one of each pair. Samples were obtained at time 0, before division of the cultures, and at 30 min, 60 min, and 120 min.
The resulting images revealed, that the inducer arabinose was required to stimulate expression of YFP, and that the addition of the repressor glucose to a uninduced culture had no effect. Furthermore, it was evident that addition of arabinose induced expression of YFP, and that subsequent addition of glucose terminated the pBAD regulated gene expression, resulting in a reduced fluorescence level. 30 minutes after inhibition this reduction was already evident, and after 120 minutes the gene expression controlled by pBAD was even further decreased, as seen in Figure 1.

Figure 1. YFP fluorescence levels in E. coli MG1655 transformed with the pBAD-YFP reporter system on pSB3K3. Left: Cultures with the inducer arabinose added. Right: Cultures not induced with arabinose. Both cultures were split up at OD600=0.1, designated time 0, and the inhibitor glucose was added to one half of each culture. Images were obtained at 0, 30, 60, and 120 minutes.
This experiment made it clear, that gene expression controlled by the pBAD promoter is both inducible and repressible as required when cloned into the low copy vector pSB3K3.
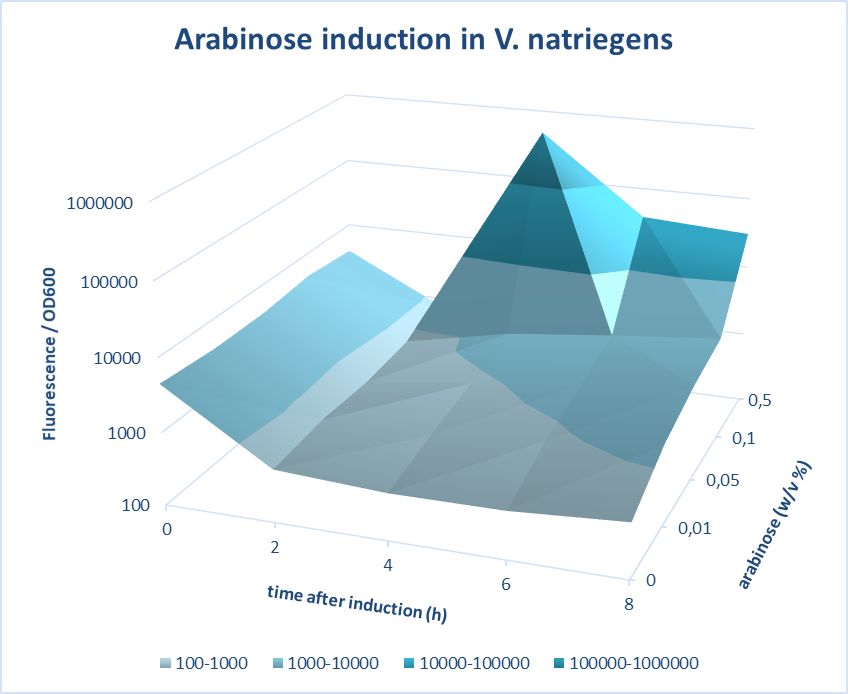
Induction of pBAD promoter in Vibrio natriegens
(Characterized by iGEM Groningen-2019)
iGEM Groningen 2019 characterized the inducibility of pBad in Vibrio natriegens. V. natriegens has been reported to have a remarkable doubling time of 9.8 min. As is the case for the better-studied but slower-growing E. coli, V. natriegens increases its number of ribosomes with the growth rate in order to achieve its extraordinarily high rate of protein synthesis. Multiple mechanisms contribute to this high ribosome synthesis efficiency, including high rRNA gene copy number; strong promoters that contain near-consensus −10, −35, and UP elements; and activation by the transcription factor Fis. In addition, V. natriegens rRNA promoters exhibit the relatively short-lived open-complex characteristic of rRNA promoters in E. coli, potentially contributing to the regulation of these promoters in vivo.
A construct of pBad-mCherry was designed using 3A assembly method. The pBad promoter was characterized at different time points after induction with different concentrations of arabinose. Further, the efficiency of pBad was measured by carrying out fluorescence measurements of the expression of mCherry on induction with different concentrations of arabinose. The screening was done based on quantifications of the fluorescence intensity measured. The best version of the promoter was determined based on fold change in the fluorescence intensity and low basal level. The optimum concentration of arabinose was found to be 0.5 w/v% at 4 hours after induction.
Figure 1. Characterization of Arabinose induction in V. natriegens.
//direction/forward
//chassis/prokaryote/ecoli
//promoter
//regulation/positive
//classic/regulatory/other
| biology | |
| control | araC, arabinose |
| direction | Forward |
| n/a | Inducible pBad/araC promoter |
| negative_regulators | |
| o_h | |
| o_l | |
| positive_regulators | 1 |

 1 Registry Star
1 Registry Star