Part:BBa_K117000:Experience
This experience page is provided so that any user may enter their experience using this part.
Please enter
how you used this part and how it worked out.
New Using Experience From iGEM2014 WHU-China
In order to secret our target protein to the extracelluar space, we added this lysis part to our project and design a series of experiments to prove the function of E7 lysis protein.
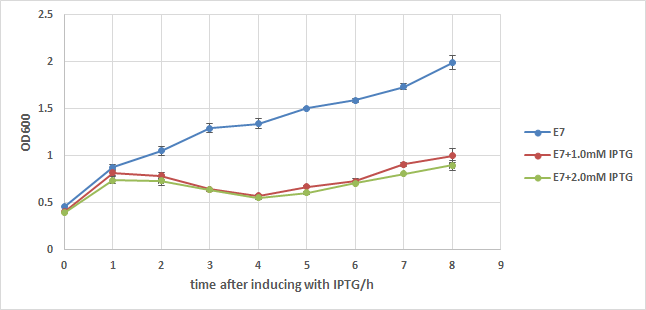
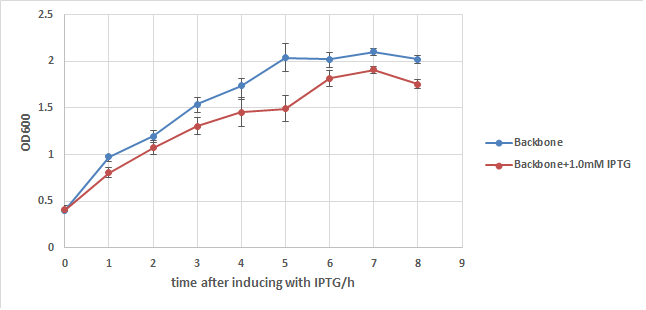
We first add a strong RBS(B0034) to this part, then integrate it to a commercialized plasmid named pUHE21, which has an IPTG inducible promoter. Next, we transform it into a strain of E.coli named w3110, which can synthesize and secret colicin to kill other type of E.coli. We also build up a negative control composed of w3110 and pUHE21 backbone. Those so called "E7 strain" and "BB(backbone) strain" are then induced with 1.0mM of IPTG at OD=0.5 after recovery. Samples are taken every hour for 8 hours and their OD are measured. The result is showed in the two charts below:
From chart1 we can see the obvious difference between induced and noninduced E7 strain 2 hours after induction, and the difference reaches about 1 fold 8 hours after induction, indicating that a large number of E.coli lyse because of the induction of E7.
In order to exclude the possibility that IPTG itself may cause drop in the growth rate of E.coli, BB strain is used to conduct another experiment. From chart2 we can see that IPTG does have some influence to the growth of E.coli, but its effect is too weak to cause such a big OD difference in chart1. Thus it's safe to say that the expression of E7 is the main cause of w3110's lysis, but it can't kill all of E.coli even under such a strong expression.
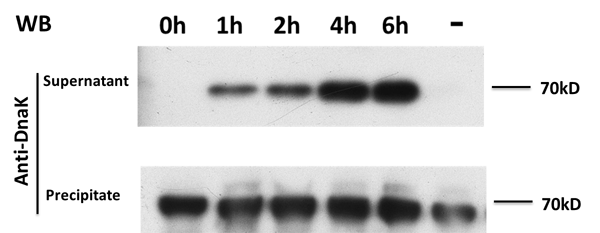
On the other hand, in order to prove that our target protein can be secret to the outside of E.coli with the help of E7, Samples are also taken every 2 hours from those groups discribed above, and after centrifugation, the supernatant and sediment are collected separately and western blot is adopted to detect a kind of endogenous protein called DnaK, which expresses itself constently in normal E.coli. The result is showed in the picture below:
From the picture we get to know that normally DnaK can't be secreted to the outside of E.coli after expression, but when E7 is expressed, E.coli lyses and DnaK leaks to the extracellular space and thus can be detected in the supernatant, and its concentration increases over time, proving the function of E7 lysis protein.
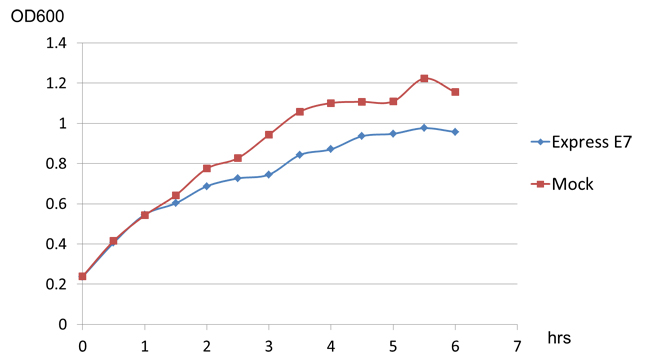
Besides those two main experiment to prove the funtion of E7 lysis protein, we also have an interesting discovery. When we transform E7 lysis gene into BL21, which can't secret colicin, we find that E7 lysis protein can still cause partially lysis in it, though the influence is not as strong as in w3110(the OD difference reaches about 0.5 after 10 hours of induction, see chart3 below).
This result indicates that E7 lysis protein may also have function in noncolicin-secreted strain of E.coli, but it remains to be explained what's the mechanism of E7 lysis protein and what causes the different effect E7 lysis have on colicin-secreted and noncolicin-secreted E.coli strain.
Applications of BBa_K117000
This Biobrick part is a Lysis gene. It can be ligated to the desired choice of promoter and lysis of the E coli cells could be acheieved.
The NTU@iGEM did just that, and ligated this biobrick part to the pLsrA promoter which detect AI2, an auto-inducer molecule. After the pLsrA detects the AI2, the promoter promotes the transcripton of the lysis gene. Next translation takes place and the lysis protein is formed.
Lysis protein would lyse the cell by the destruction of the cell membrane.
For more detail description of our the Lysis part was used by the NTU@iGEM team, please visit: [http://2008.igem.org/Team:NTU-Singapore NTU@iGEM Team Main Page].
Using experience 2
we seu_a used and improved this part,details are as follow:
This part we use R0010 promoter ,it is to say in the absence of LacI protein and CAP protein, it promotes transcription,in the presence of LacI protein and CAP protein, it inhibits transcription. When induced by IPTG, the protein expression begin.
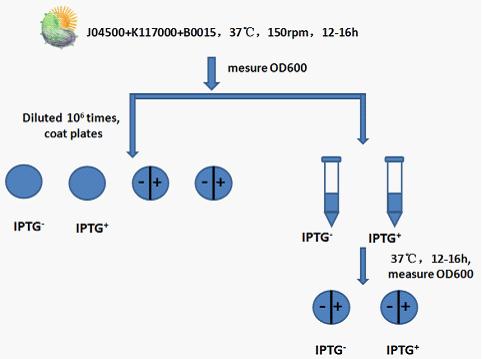
Experimental procedure:
Step 1: Recovery J04500+K117000+B0015,shaking it in 37℃ for 12 to 16h at 150rpm.
Step 2: Measure the OD600 of bacterial liquid.
Step 3: Use 50ul bacterial liquid which has been diluted times to coat plates.Set IPTG(none or 1mM) control.
Step 4: Add 5ul bacterial liquid which has been diluted times to 5ml liquid Luria-Bertani medium. Set IPTG(none or 1mM) control. After cultivate in 37℃ shaking(150rmp) for 12 to 16h,measure the OD600 of bacterial liquid .
Step 5: Use 50ul bacterial liquid(from step 4) which has been diluted times to coat plates. Set IPTG(none or 1mM) control.
Step 6: Compare bacterial liquid concentration and petri dishes on the growth of the colony to judge lethal gene’s effects .Compare the bacterial concentration and Petri dish colony growth to judge the effect of lethal gene. Judging from the OD600 of bacterial liquid and the growth state of bacteria in Petri dish to see whether the death gene work or not.
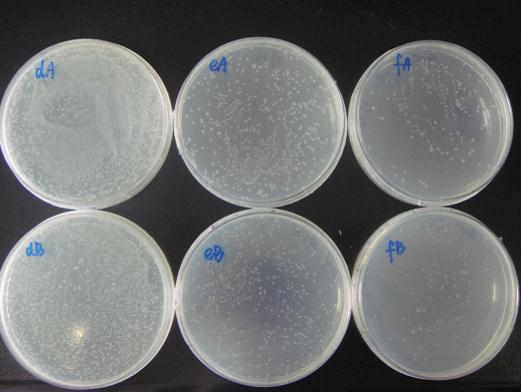
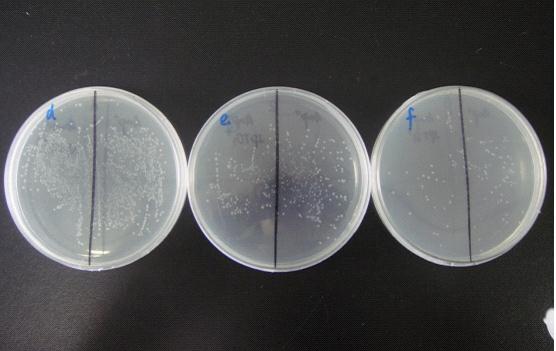
(Notes: d/e/f means the bacterial liquid were diluted 104/105/106times,A/B means the concentration of IPTG none/1mM )
The growth state of bacteria in Petri dish:
The comparation of colony growth:
We can see from the figure that the bacteria caring the death gene is in bad condition comprared to the one without . We conclude that the death genes work.
For more information on how this part operates in our system, please visit [http://2012.igem.org/Team:SEU_A SEU_a Team Main Page].
New Using Experience From SZU-China 2019 iGEM
With the purpose of using a biological cell cleavage method instead of the traditional physical methods, our group aim to create a mechanism with a special function which is able to break the host cell's structure. Besides, the whole lysing metabolism should be stable and controllable. And fortunately, cell lysis metabolism triggered by lysis protein is very stable and is widely used by LMB(Laboratory of Molecular Biology). As a result, to meet our need, we just need to simply add a Tryptophan attenuator before the part BBa_K117000.
SZU-China 2019 iGEM team constructed a plasmid with BBa_K1475900 (Promoter), BBa_K2912014 (Attenuator), BBa_K117000(lysis), BBa_K3257020(terminator).
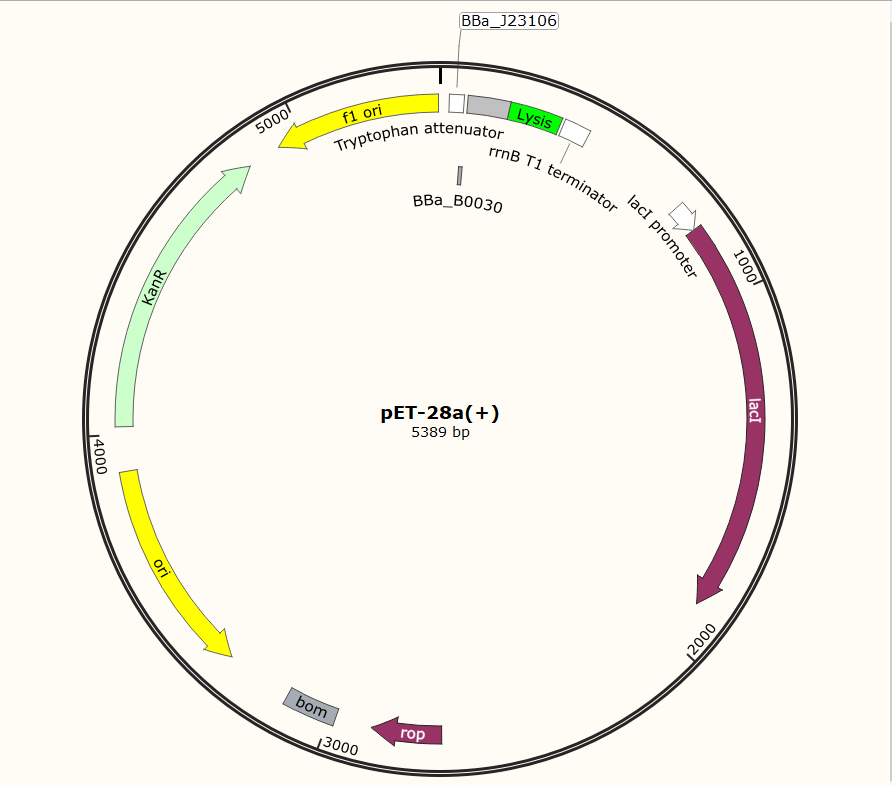
Before the official experiment of Trp_Lysis gene. We have done the T7 promotor_lysis gene characterization experiment with IPTG inducing in LB liquid medium(illustrated with Fig.2).

According to the characterization experiment's result, we can draw a conclusion that once the lysis protein is produced with IPTG inducing by HT115 (DE3) E.coli, their growth will be significantly inhibited.
In our next step, we transformed the expression vector into HT115 (DE3) E.coli by heat-shock method, culturing in LB liquid medium to exact exponential phase, and then moved to fresh LB liquid medium with different concentration of tryptophan, starting a eight-hour cell growth curve experiment of our engineering E.coli and then we measured the OD600 of each groups every two hours. Finally, we can calculated their survival rate by use OD600 of the experimental group divided by OD600 of the blank group (illustrated with Fig.3).

Besides, we also did a culturing in LB solid medium experiment in tryptophan concentration gradient.(illustrated with Fig.4)
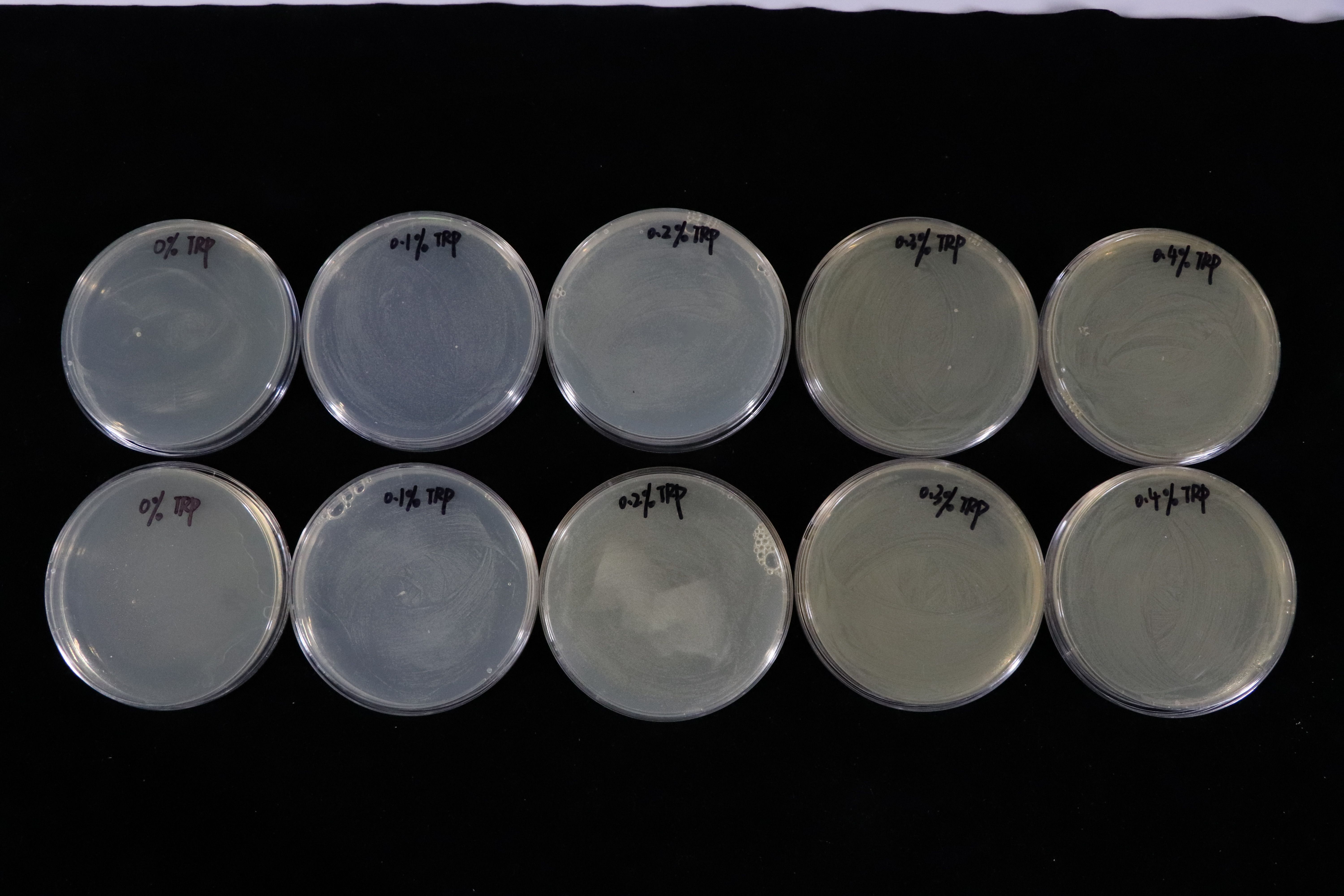

In contrast with T7 promotor_lysis with IPTG inducing group(illustrated with Fig.5). The Trp_Lysis metabolism obviously has the advantage which is able to kill the host cell by breaking their structure in a controlled way. In other words, we can switch off or switch on the lysing mechanism by controlling the tryptophan concentration of the culture medium, and the result indicates that the concentration critical valve of tryptophan is around 0.3% which means that if the concentration is more than 0.3%, the lysing mechanism will be closed so that the host cells can grow as usual.
User Reviews
UNIQe413b07c180b7840-partinfo-00000005-QINU UNIQe413b07c180b7840-partinfo-00000006-QINU

 1 Registry Star
1 Registry Star