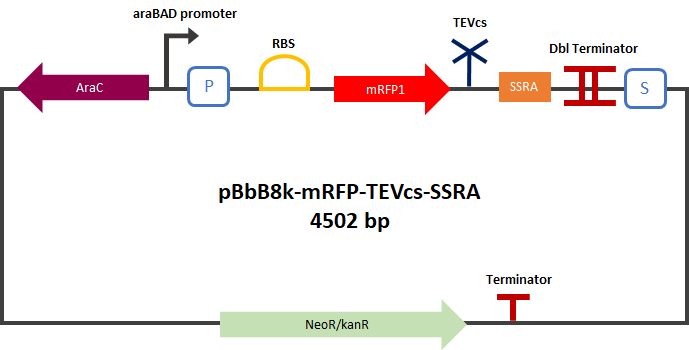
Part:BBa_K3147003
mRFP1 fused to a TEV-cleavable ssrA tag
I : parts BBa_K3147003 (mRFP1-TEVcs-SSRA) function
The Montpellier 2019 team made a reporter gene construction in order to carry out their proof of concept. This construction produces an mRFP1 (BBA_E1010) fused in C-ter with a fast degradation tag called ssrA [2] (BBA_M0050). The TEV cutting site (BBa_J18918) was added between the mRFP1 and the ssrA tag. This construction can be used as a reporter gene. In the presence of TEV the ssrA is separated and allows the mRFP to glow without being degraded.
II. Proof of function
The experimental approach to test the activity of these reporters was to compare the basal fluorescence rate of mRFP1 with a TEV cutting site "cleaved" to this construction. For this purpose we made a control construction: mRFP1-TEVcs (BBa_K3147004) similar to this one by removing the proteolysis tag and simulating a cut by the TEV. The construction was cloned by Gibson Assembly in a pBbB8k-GFP (https://www.addgene.org/35363/ ) backbone under the control of a BAD type promoter, in order to control its expression.
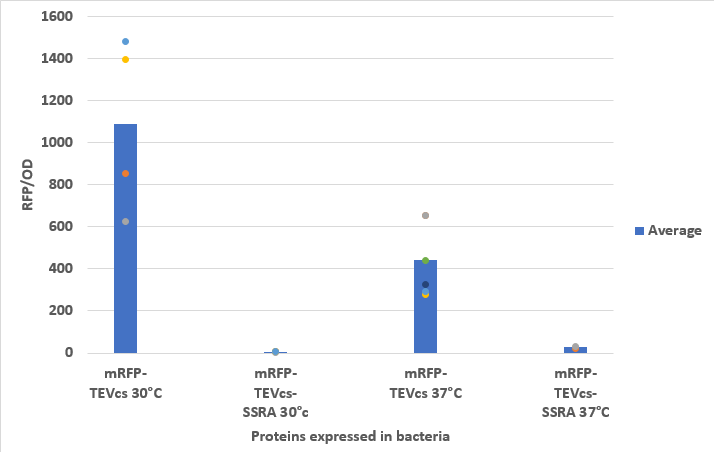
We compared the basal fluorescence of strain E. coli NEB10β transformed with the mRFP1-TEVcs construction to an E. coli NEB10β strain transformed with the mRFP1-TEVcs-ssrA construction. Fluorescence was quantified after induction with arabinose concentrated at 1% with the plate reader overnight. Here are the fluorescence of mRFP-TEVcs-ssrA and mRFP-TEVcs at 30°C and 37°C. We can see that the ssrA tag is reducing the fluorescence from the mRFP-TEVcs.
Reference
[2] Sunohara, T., Abo, T., Inada, T., & Aiba, H. (2002). The C-terminal amino acid sequence of nascent peptide is a major determinant of SsrA tagging at all three stop codons. RNA (New York, N.Y.), 8(11), 1416–1427. doi:10.1017/s1355838202020198
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25INCOMPATIBLE WITH RFC[25]Illegal NgoMIV site found at 691
- 1000COMPATIBLE WITH RFC[1000]
| None |