Part:BBa_I746909:Experience
This experience page is provided so that any user may enter their experience using this part.
Please enter
how you used this part and how it worked out.
Applications of BBa_I746909
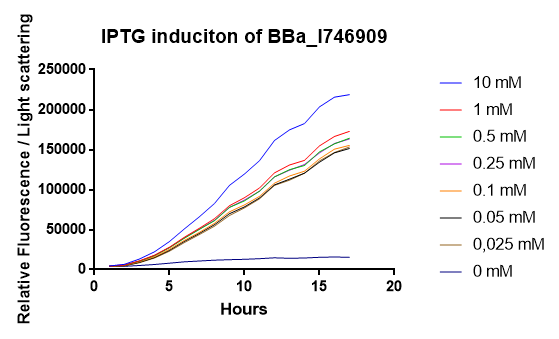
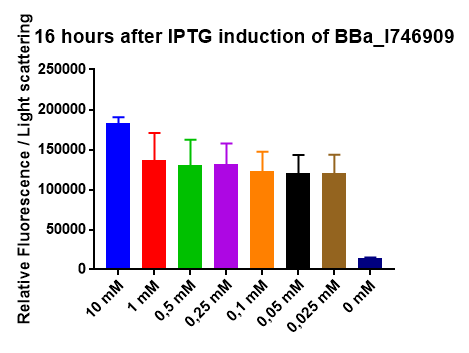
2018 iGEM team Linkoping Sweden
2018 iGEM team Linkoping Sweden validated this part.
[http://2014.igem.org/Team:Imperial iGEM Team Imperial
[http://2014.igem.org/Team:Aachen iGEM Team Aachen
The iGEM Team Aachen used the biobrick I746909 to test their [http://2014.igem.org/Team:Aachen/Project/2D_Biosensor sensor chip technology].

|
| Testing I746909 in sensor chips I746909 in sensor chips induced with 2µl IPTG and measured with a plate reader. Top chip is not induced, bottom chip is induced with IPTG. |
The biorbick I746909 was also used to test the measurement device [http://2014.igem.org/Team:Aachen/Project/Measurement_Device WatsOn].

|
| Testing I746909 in WatsOn I746909 in sensor chips induced with 2µl IPTG and measured with WatsOn. Left chip is not induced, right chip is induced with IPTG. |
2015 iGEM team Bielefeld-CeBiTec
2015 iGEM team Bielefeld-CeBiTec used this part and improved it by addition of a designed, translation enhancing 5'-untranslated region (5'-UTR; BBa_K1758100). You can find further information at BBa_K1758102).
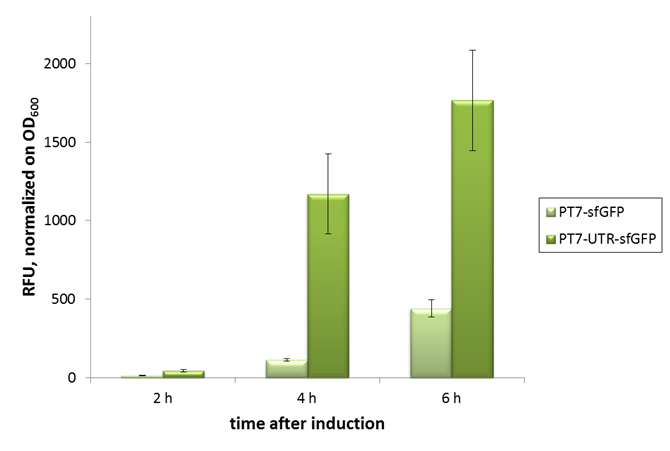
As can be seen in the picture, the difference was observable with the naked eye as well.
[http://2014.igem.org/Team:Imperial iGEM Team Imperial
[http://2014.igem.org/Team:Aachen iGEM Team Aachen
The iGEM Team Aachen used the biobrick I746909 to test their [http://2014.igem.org/Team:Aachen/Project/2D_Biosensor sensor chip technology].

|
| Testing I746909 in sensor chips I746909 in sensor chips induced with 2µl IPTG and measured with a plate reader. Top chip is not induced, bottom chip is induced with IPTG. |
The biorbick I746909 was also used to test the measurement device [http://2014.igem.org/Team:Aachen/Project/Measurement_Device WatsOn].

|
| Testing I746909 in WatsOn I746909 in sensor chips induced with 2µl IPTG and measured with WatsOn. Left chip is not induced, right chip is induced with IPTG. |
User Reviews
UNIQ658b3d1b2a3def59-partinfo-00000006-QINU UNIQ658b3d1b2a3def59-partinfo-00000007-QINU

 1 Registry Star
1 Registry Star

