Part:BBa_K2003011
Inducible Green Fluorescent Protein UnaG+6xHis-tag+Flexible linker
UnaG was discovered as late as 2013. It originates from the muscle fibers of the Japanese Unagi eel Anguilla japonica. This is the first fluorescent protein to be found in a vertebrate species, giving it a range of biochemical properties that differs from proteins found in the original FP family - one of the most astounding ones being that fluorescence occurs only when a ligand binds non-covalently to the protein, namely bilirubin.
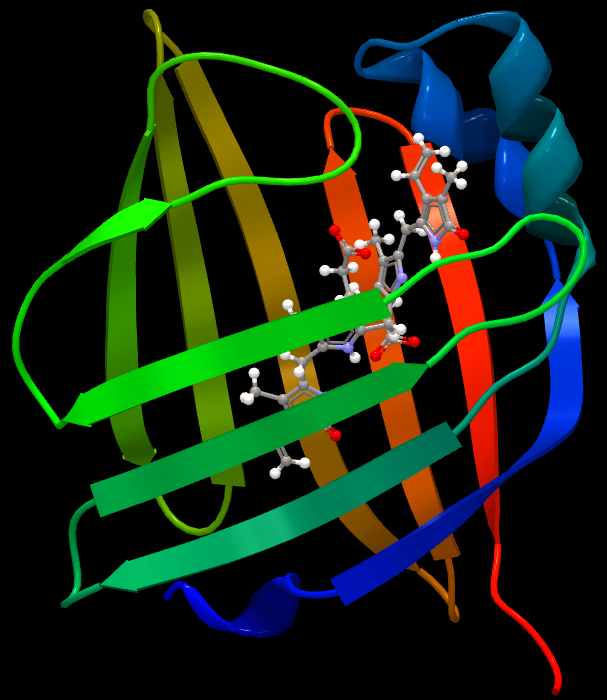
Structure of UnaGUnaG is a fatty acid-binding protein (FABP). These proteins are cytosolic and exist in most vertebrates and invertebrates. They contribute to the transport of lipophilic compounds, often fatty acids. Although it is still rather evasive in which ways FABPs function in cells, some of these proteins appear to play roles in cellular anabolism. Wild-type UnaG is 139 amino acids long and has a molecular weight of 15.6 kDa. UnaG is a ß barrel structure containing eleven ß strands - a typical quality for FABPs (Figure 1). Instead of binding a fatty acid molecule, the ligand of UnaG is bilirubin, which binds non-covalently to the center of this ß barrel. Bilirubin exists in all vertebrates and is a metabolite of the heme group in hemoglobin. Bilirubin can only bind to UnaG in its unconjugated (4Z,15Z) form, meaning that wild-type UnaG shows high specificity to this particular molecule. In fact, BR is the actual fluorophore of UnaG. When BR binds to the cavity of the protein, it emits green light at 527 nm. Holo-UnaG has maximum absorption at 498 (i.e. blue light) nm. UnaG does not need molecular oxygen in order to fluoresce, making it suitable for research executed under anaerobic conditions where FPs are not functional. According to Akiko Kumagai and his team, the quantum yield of holo-UnaG is 0.51 and it is stable in a pH ranging from 4 to 11. GFPs are usually stable in a pH range from 6-10, but this varies depending on which mutant is used.
Using UnaG as a Fluorescent MarkerApo-UnaG does not fluoresce, but Akiko Kumagai’s research group showed that Holo-UnaG (i.e. UnaG+BR) fluoresces instantaneously. No other fluorescent protein has a ligand that binds non-covalently to it. This could mean that it is possible to create an inducible fluorescent marker that could be used instead of photoswitchable FPs. Bilirubin does not exist naturally in lower classes of organisms such as bacteria and yeast, so it would have to be added exogenously when research is being done on these types of cells. UnaG has shown the ability of being fused to other proteins via a flexible linker both on the C terminus and the N terminus, making it suitable to use as a fusion marker.
Since UnaG was discovered fairly recently, no mutagenic variations of the protein have been developed with improved characteristics than those of wtUnaG. However, several fluorescent FABPs have recently been discovered in other marine organisms. Two of them are called Chlopsid FPI and FPII. These two were also found in eels, although in another species, and both of them emit green fluorescence. Researchers have identified a tripeptide motif – Gly-Pro-Pro – conserved in all fluorescent FABPs. Chlopsid FPI and FPII are also bilirubin inducible. Alignment of the DNA sequences of UnaG and the Chlopsid proteins show 54% homology. There is a slight blue shift in the fluorescence (UnaG is 498/527 nm ex/em and Chlopsid FPI and FPII 489/523 nm ex/em). The researchers suggest that this blue shift stems from the fact that the 57th amino acid in UnaG is asparagine, whereas Chlopsid FPI and FPII contain a histidine at this site. The histidine would contribute to a more π-conjugated system, resulting in a difference in fluorescence. This discovery suggests possibilities of changing the emission wavelength of these proteins by mutagenesis comparableto what has been done on GFP for the past 40 years. This could in that case enable creation of fluorescent FABPs of different colors.
Note that Uppsala 2018 performed a correction on this part by moving a faulty start codon. More information can be found at the [[Part:BBa_K2669001]] page.
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000COMPATIBLE WITH RFC[1000]
| None |