Part:BBa_K2637011
TDH3 Promoter
TDH3 promoter is a strong yeast based promoter of glyceraldehyde -3- phosphate dehydrogenase, also known as GDS or GAPDH, which is often used for expression of exogenous genes in yeast.
Our basic parts range from BBa_K2637001 to BBa_K2637018 and BBa_K2637053 to BBa_K2637059. Our composite parts range from BBa_K2637021 to BBa_K2637044 and BBa_K2637047 to BBa_K2637052. Our part collection include all our basic parts except for BBa_K2637018 and all composite parts.
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21INCOMPATIBLE WITH RFC[21]Illegal BglII site found at 366
- 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000COMPATIBLE WITH RFC[1000]
Contents
Characterization of Life Tik Tok
Characterization of promoters
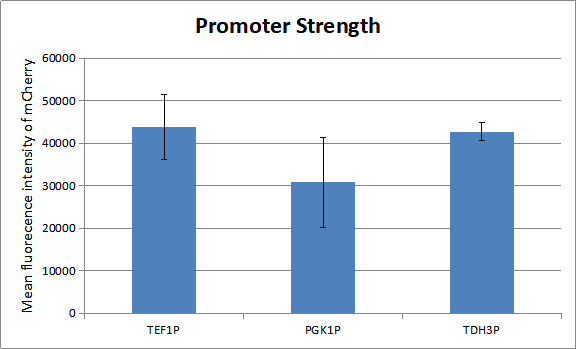
Before our test the whole system, we measured the strength of promoters which regulate the experssion of genes in our oscillator. As there is a natural ratio of the three proteins-KaiA, KaiB and KaiC in cyanobacteria. And a certain ratio of the three proteins has advantage of modeling.
We can draw a conclution about the strength of the three promoters. So we combine TEF1 promoter with KaiA and TEF1 terminator, PGK1 promoter with KaiB and PGK1 terminator and TDH3 promoter with AD-KaiC and ADH1 terminator. Similarly, we combina TEF1 promoter with CikA and TEF1 terminator, PGK1 promoter with RpaA and PGK1 terminator and TDH3 promoter with BD-SasA and ADH1 terminator. And here is only one combination of our experiment. To get more combinations, please see the part from BBa_K2637027 to BBa_K2637038.You can see more about our project to click here.[http://2018.igem.org/Team:Tianjin]
Characterization of Life Tik Tok
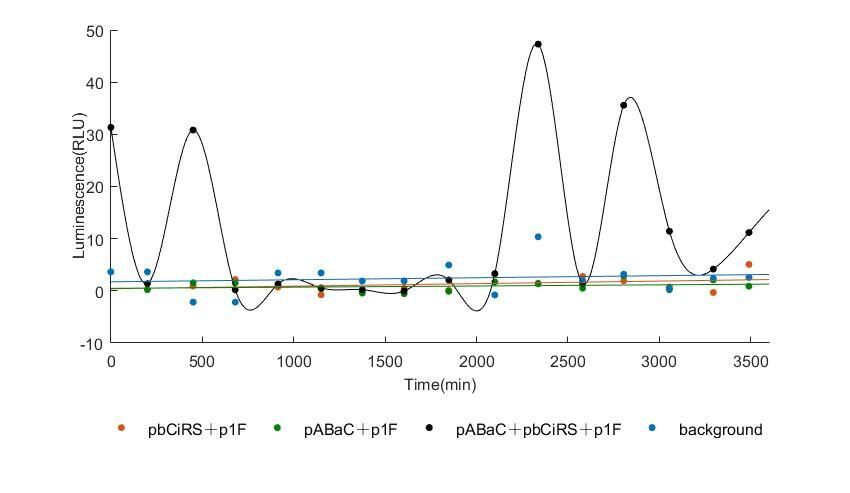
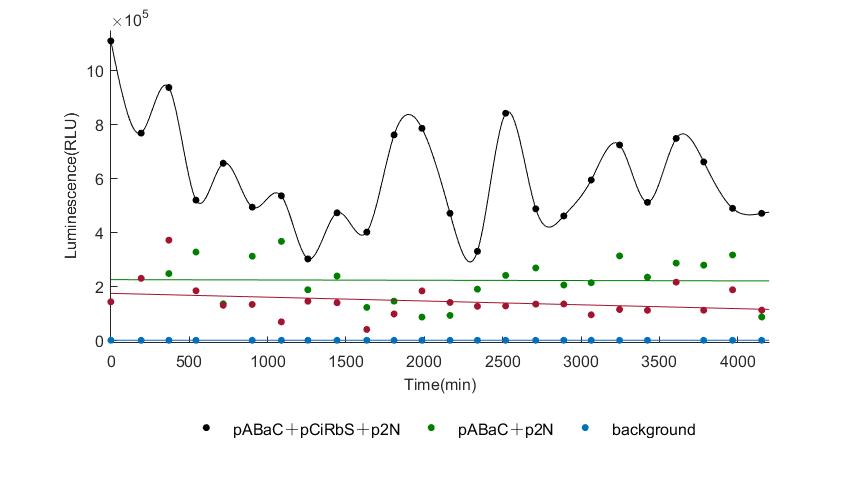
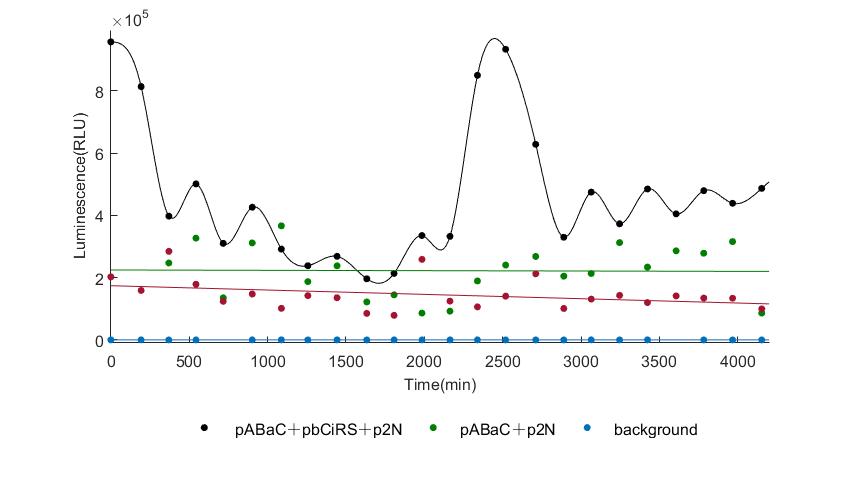
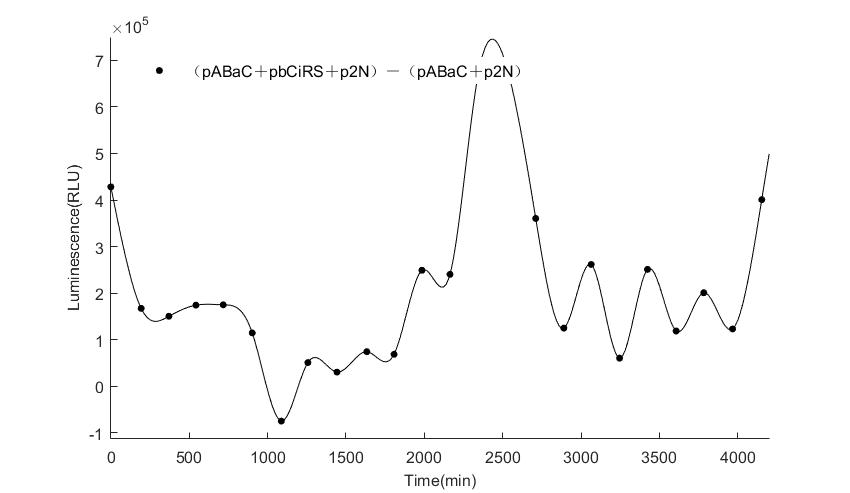
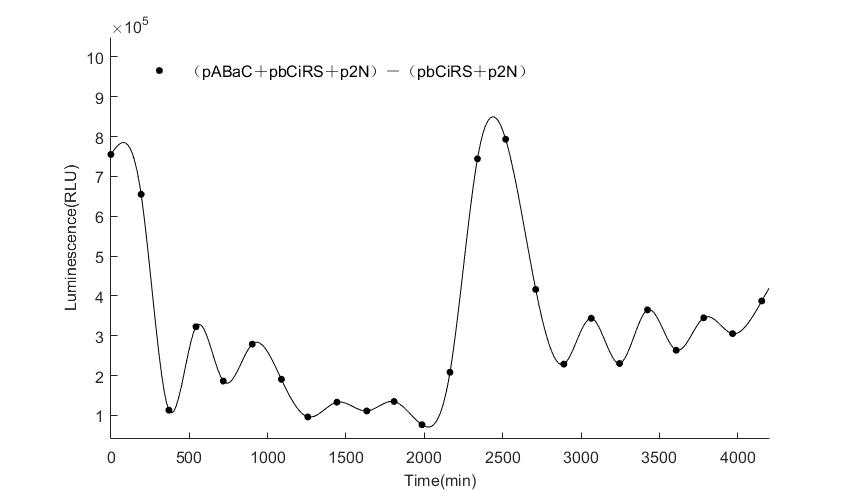
When we start to measure the datas of experiment group, we set up some control groups at the same time. To prove that our oscillator works surely because of the yeast two-hybrid system, we constructed some bacteria in which there are only one fusion protein of yeast two-hybrid system and a reporter gene or a combination. And we choose 3 or 4 reporter genes as parallel experiment groups.
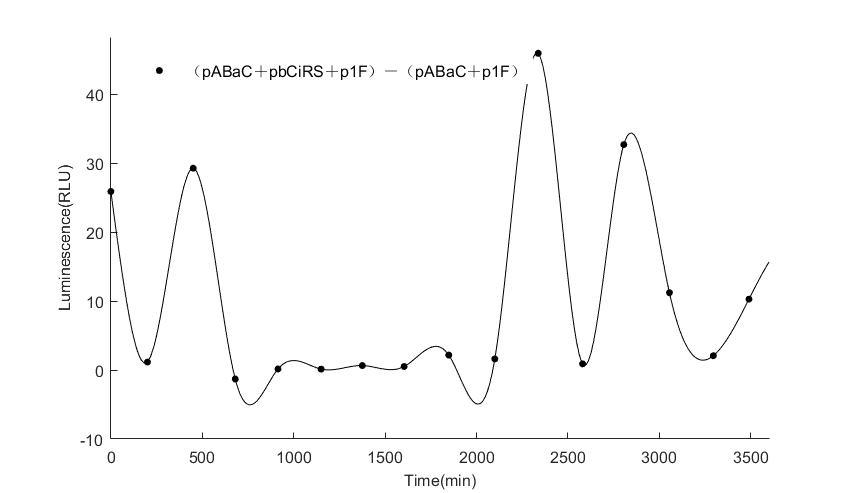
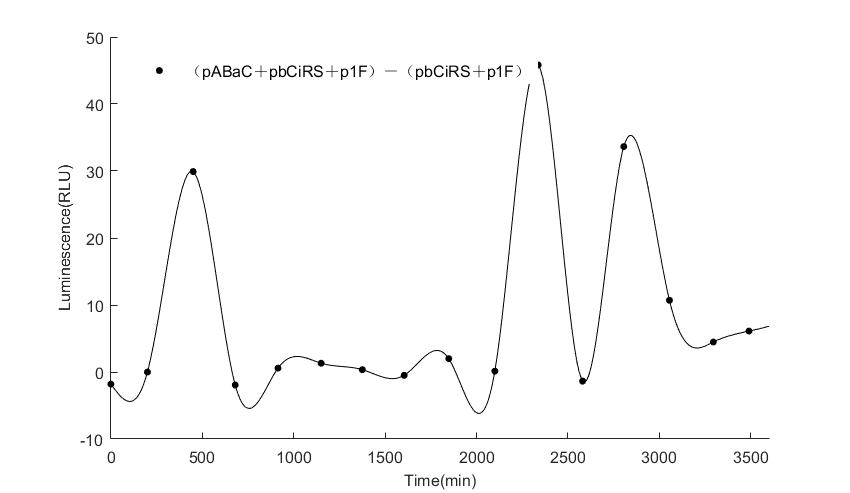
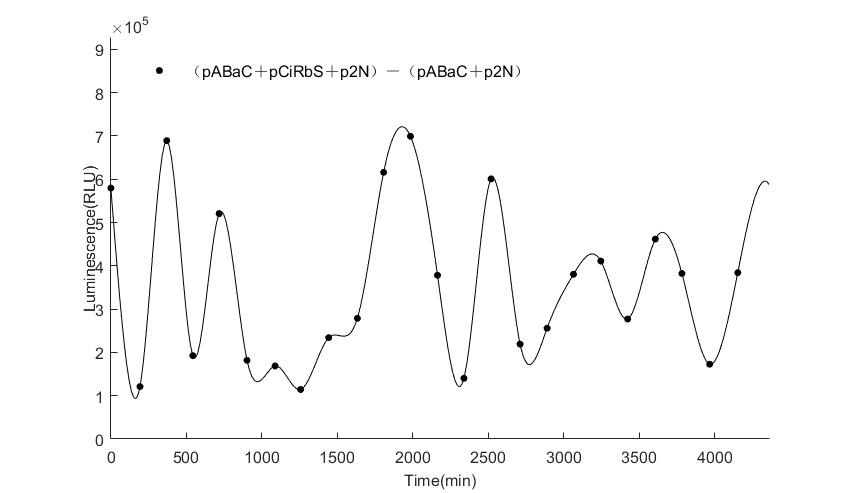
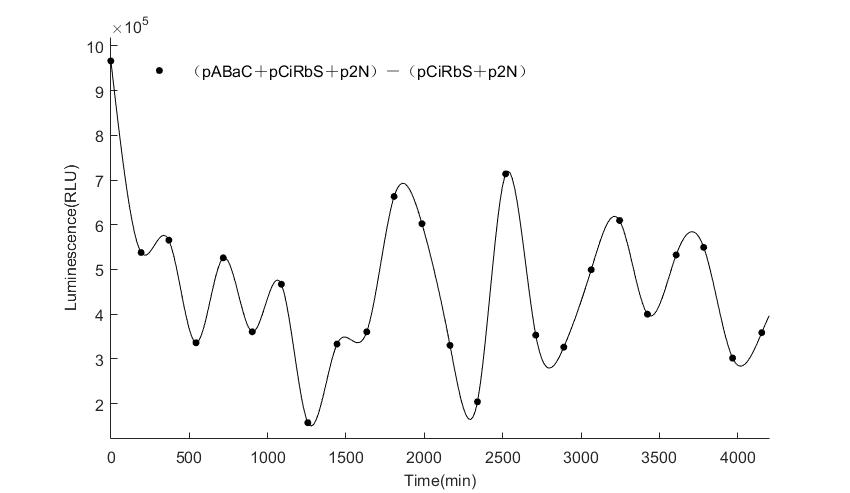
In the above figures, pABaC is the plasmid with AD-KaiC fused gene and make up a yeast two-hybrid system with the plasmid pbCiRS, which is with BD-CikA fused gene. p1F is the plasmid with Fluc reporter whose promoter is Gal1. In the 1st figure, the experimental group have black datas and the first control group and the second group have green and red datas, respectively. We substracted the control groups to gat a more objective result. As we predicted and looked at the results in other literatures, this oscillatory system is unstable.However, even though the fluctuations are not obvious on the second day, better waveforms could be observed on the first and third days.In addition, on the third day we get very high peak values, and in the wave trough experimental report gene expression is closer to the control group and the blank group, this indicates that in subjective day, yeast two-hybrid system works and results the peak value, in subjective night, it doesn't work and causes the similar results to the control groups and the background. Therefore, the overall experimental results show that we successfully reconstructed the oscillator in yeast.
And the following figures are the results we measured with live Nanoluc. We get the similar conclusion to the Fluc group.
You can see more details of our results [http://2018.igem.org/Team:Tianjin/Demonstrate here]
References
[1]Roger Tseng, Nicolette F. Goularte, Archana Chavan, Jansen Luu, Susan E. Cohen, Yong-Gang Chang, Joel Heisler, Sheng Li, Alicia K. Michael, Sarvind Tripathi, Susan S. Golden, Andy LiWang, Carrie L. Partch, Structural basis of the day-night transition in a bacterial circadian clock. Science, 1174-1180 (2017).
[2]Joost Snijder, Jan M. Schuller, Anika Wiegard, Philip Lössl, Nicolas Schmelling, Ilka M. Axmann, Jürgen M. Plitzko, Friedrich Förster, Albert J. R. Heck, Structures of the cyanobacterial circadian oscillator frozen in a fully assembled state. Science, 1181-1184 (2017).
[3]Joseph S.Markson, Joseph R.Piechura, Anna M.Puszynska, Erin K.O’Shea, Circadian Control of Global Gene Expression by the Cyanobacterial Master Regulator RpaA. Cell, 1396-1408 (2013).
[4]Jun O. Liu,et al.Everything you need to know about the yeast two-hybrid system.DOI:10.1038/788
[5]Hideo Iwasaki,* Stanly B. Williams,et al.A KaiC-Interacting Sensory Histidine Kinase, SasA, Necessary to Sustain Robust Circadian Oscillation in Cyanobacteria.Volume 101, Issue 2
//promoter
| biology | S. cerevisiae |
| chassis | S. cerevisiae |