Part:BBa_K2541401
sfGFP(BbsI free)
It is a superfolder GFP, a robustly folded version of GFP, which is BbsI restriction site free and can be used in GoldenGate assembly.
1. Usage and Biology
Green fluorescent protein (GFP) exhibits intrinsic fluorescence and is commonly used as a reporter gene in intact cells and organisms [1]. Many mutants of the protein with either modified spectral properties, increased fluorescence intensity, or improved folding properties have been reported [2].
GFP often misfolds when expressed as fusions with other proteins, while a robustly folded version of GFP, called superfolder GFP (sfGFP), was developed and described by Pédelacq et al at 2006[3] that folds well even when fused to poorly folded polypeptides. We decided to use sfGFP as our reporter protein due to its faster folded feature and higher fluorescence intensity. The superfolder GFP had been registered in iGEM BBa_I746916.
However, the superfolder GFP (BBa_I746916) contains a BbsI restriction enzyme cleavage site, and the BbsI restriction endonuclease is an economical and efficient enzyme used in GoldenGate assembly, so the superfolder GFP (BBa_I746916) cannot be used for GoldenGate assembly. This year we made two point mutations in superfolder GFP (BBa_I746916) to eliminate its BbsI restriction site without changing its amino acid sequence to make it suitable for GoldenGate assembly. We changed the sequence from 5’-gaagac-3’ to 5’-gaggat-3’. And we called the mutated variant sfGFP(BbsI free) (BBa_K2541401).
Figure 1. BbsI restriction site and point mutation.
Figure 2. Expression of three types of sfGFP(BBa_I746916, BBa_K2541401, BBa_K2541400), cultivated overnight.
2. Characterization
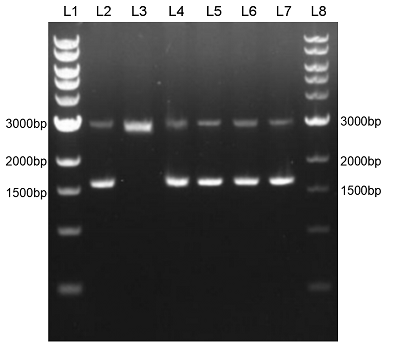
We constructed sfGFP(BbsI free) (BBa_K2541401) sequence and superfolder GFP (BBa_I746916) on the pSB1C3 vector. The BbsI recognition site free was confirmed by nucleic acid electrophoresis (figure 2). The sfGFP(BbsI free) sequence can not be digested by BbsI. And the length of these sequences are correct.
Figure 3. L1: 1kb DNA marker; L2: BBa_I746916; L3: BBa_I746916+BbsI; L4: BBa_K2541401; L5: BBa_K2541401+BbsI; L6: BBa_K2541400; L7: BBa_K2541400+BbsI; L8: 1kb DNA marker.
We got the emission and excitation spectra of two type sfGFP: sfGFP(BbsI free) (BBa_K2541401) and superfolder GFP (BBa_I746916) (figure 2).
Figure 4.
For further characterization, we detected the expression intensity of these two types of sfGFP. According to the results (figure 3), we found out that the fluorescence intensity of sfGFP(BbsI free) (BBa_K2541401) is higher than superfolder GFP (BBa_I746916).
Figure 5. The expression of three types of sfGFP in E.coli. The grey line represents fluorescent expression of sfGFP_optimism (BBa_K2541400), the orange line represents fluorescent expression of sfGFP(BbsI free) (BBa_K2541401), the blue line represents fluorescent expression of superfolder GFP (BBa_I746916) and the yellow line represents fluorescent expression of negtive control (BBa_J364007).
3. Conclusion
We have made an improvement on the superfolder GFP (BBa_I746916). Our sfGFP(BbsI free) (BBa_K2541401) is BbsI restriction site free, which can be used in GoldenGate assembly to achieve efficient and rapid assembly of gene fragments. And its fluorescence intensity is higher than superfolder GFP (BBa_I746916).
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000INCOMPATIBLE WITH RFC[1000]Illegal SapI.rc site found at 13
| None |