Part:BBa_K2601002
SUMO (Small Ubiquitin-like Modifier)
Introduction
SUMOylation is a post-translational modification, which is involved in many cellular processes and provide a versatile way to regulate the dynamics of protein-protein interactions. Conjugation of the ubiquitin-related SUMO modifier to target proteins provides a platform for protein-protein interactions and ordered assembly of multi-protein complexes. In humans, three SUMO forms (SUMO1, SUMO2, and SUMO3) can be attached to lysine residues of target proteins. Modification of proteins by SUMO are recognized by SUMO-interacting motifs termed SIMs. In natural process, polySUMOylation recruits distinct interaction partners, such as E3 ubiquitin ligases, that bind to polySUMO chains through tandem SIMs. SIMs bind to a surface patch between the α-helix and a β-sheet of the SUMO protein and extend the β-sheet of SUMO by one additional strand. the SIM either attaches as a parallel or an antiparallel strand to the SUMO β-sheet. Binding is primarily mediated by a stretch of four residues containing 3–4 hydrophobic amino acids (I, V, or L). This core interaction motif is a common property of all SIMs.
Design
Some membrane-less organelles, such as stress granules and P bodies, have been discovered in recent years. Proteins condense into droplets and assemble these organelles through a process called phase separation. Physically, phase separation is the transformation of a thermodynamic system from one phase to another, much like how oil and water demix from each other. According to thermodynamics, material will diffuse down the gradient of chemical potential instead of concentration. This is exactly why proteins will self organize into granules, diffusing from regions of low concentration to regions of high concentration. Here is an illustration of phase separation in cells.
 | 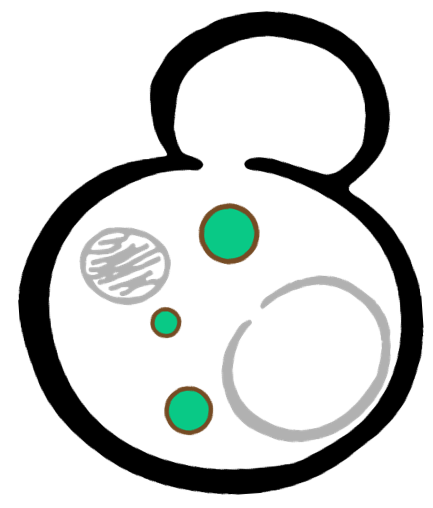 |
In order to rationally design a synthetic organelle based on protein phase separation, we need a multivalent module and a protein-protein interaction module. The paired SUMO and SIM is one of the bioparts that we chose to introduce protein-protein interaction. SUMO and SIM can dimerize spontaneously. As for the multivalent module, we turned to de novo-designed homo-oligomeric short peptides. These short peptides are called HO-Tags (homo-oligomeric tags). HOTags contain approximately 30 amino acids. HOTags have high stoichiometry, forming hexamer or tetrameric spontaneously. The hexameric HOTag3, together with the tetrameric HOTag6, can robustly drive protein phase separation upon protein interaction (achieved by SUMO-SIM module). To verify the feasibility of the system, we fused two fluorescence proteins with the two components of synthetic organelles. Thus, we can observe the self-organization of components and the formation of organelles under fluorescence microscope. Here is a demonstration of our overall design.
Properties
Previous studies reveal that the electrostatic interactions can be modulated through PTMs on either SIM or SUMO, thereby regulating the dynamics and specificity of their interactions. Reversible phosphorylation/dephosphorylation of the serine or threonine residues in phosphoSIMs dramatically enhances the affinity of SUMO binding. Biophysical measurements of selected interactions revealed a phospho-dependent shift of the dissociation constant Kd from around 50 to 1.5 μM. The specific SUMO and SIM in our design were previously characterized in Michael K. Rosen’s lab. In their studies, the apparent polySUMO-polySIM dissociation constant Kd is 70–10,000 nM, estimated from ITC measurements with (SUMO)m + (SIM)m (m = 1, 2 or 3).
Our results confirm that SUMO/SIM module fused with HOTags can drive phase separation in yeast.
Furthermore, we used Tet07, an inducible promoter, to control the expression of the SIM component. SIM fused with HOTag6 can’t express in the absence of doxycyclin, which is the activator of the promoter. Before we add doxycyclin, SUMO-HOtag3 was evenly distributed in the cells and can’t phase separate. But after we add doxycyclin, phase separation gradually appears. Therefore, we confirm the dimerization modules is essential for phase separation and HOTags alone can’t induce the process.
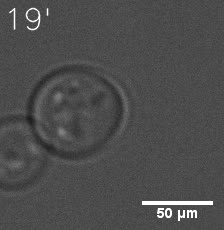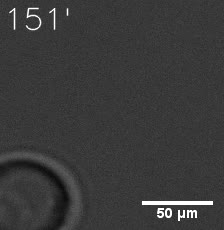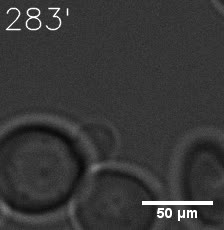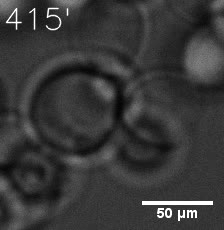 | 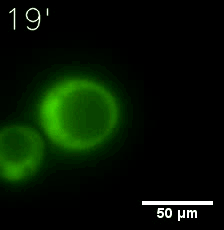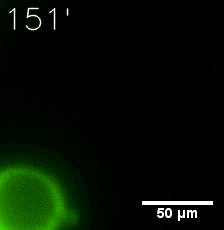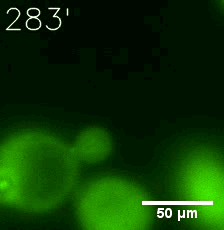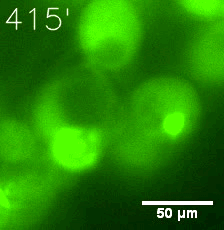 |
 | 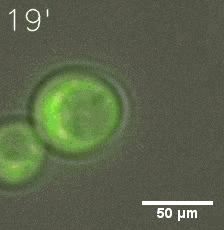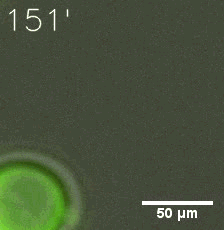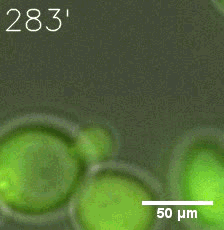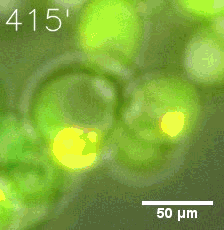 |
| Figure 4. | |
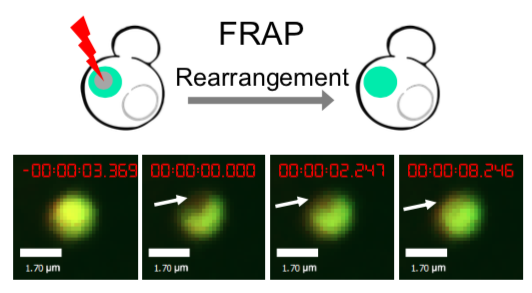
As for the biophysical properties of the system, we assume the granules are liquid-like. A liquid-like granule is in a more dynamic state, exchanging mass with cytoplasm frequently and responding to environment rapidly. There are three characteristics which define a liquid-like compartment. First, the compartments should be roughly spherical due to surface tension. It was proved by the 3D-rendered shape of the granules, taken by confocal microscope. The picture showed that the granules are nearly spherical as the aspect rate (ratio between the long axis and short axis of the synthetic granule) is about xx. Second, two droplets should fuse and coalesce into one droplet. We can see from the GIFs that the synthetic system meet this criteria. Third, the components should undergo rapid internal rearrangement. We used FRAP to verify rapid rearrangement inside the granule. After photobleaching, the fluorescence of granules quickly recovered, which indicated that the granules had rapid mass exchange with the cytoplasm.
 | 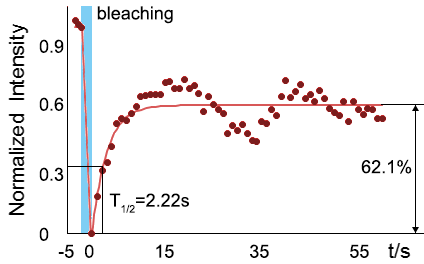 |
| Figure 6.The 3D-rendered image of granules formed by SUMO-HOtag3/ SIM-HOtag6 and the recovery of SUMO-SIM mediated organelle after fluorescence bleaching. | |
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000COMPATIBLE WITH RFC[1000]
//chassis/eukaryote/yeast
//proteindomain/binding
| biology | Saccharomyces cerevisiae |