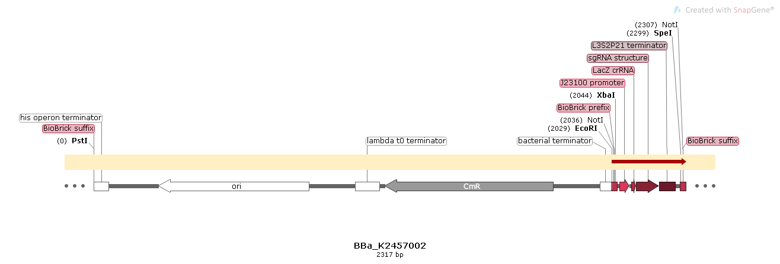
Part:BBa_K2457002
Optimized sgRNA targeting LacZ
BBa_K2457002 was engineered to be a building block of the standard BioBrick toolbox for bacterial genome: the ([http://2017.igem.org/Team:Amazonas_Brazil pCRISPeasy]). It’s the first BioBrick available on Registry with an improved sgRNA sequence - both crRNA design and tracrRNA secondary structure - to increase the CRISPR-Cas9 editing efficiency. BBa_K2457002 is composed by: the J23100 constitutive promoter sequence + an optimized sgRNA (crRNA with a GG motif at the 3’ end of it’s target-specific sequence from LacZ [1] and tracrRNA with extended duplex length and a mutation at the thymine 4 [2]) + L3S2P21 terminator [3]. It was rationally designed to be standardized AND interchangeable, aiming to provide a BioBrick to attend multiple genome editing purposes. For that matter, It’s easy to engineer new modules aiming to insert information into the genome through RFC10 Assembly.
Usage and Biology
Single-guide RNA (sgRNA) is one of the two building blocks of the CRISPR-Cas9 genome editing machinery, which directs the Cas9 protein to introduce double-stranded breaks (DSB) into target genomes sequences.
Design
Characterization
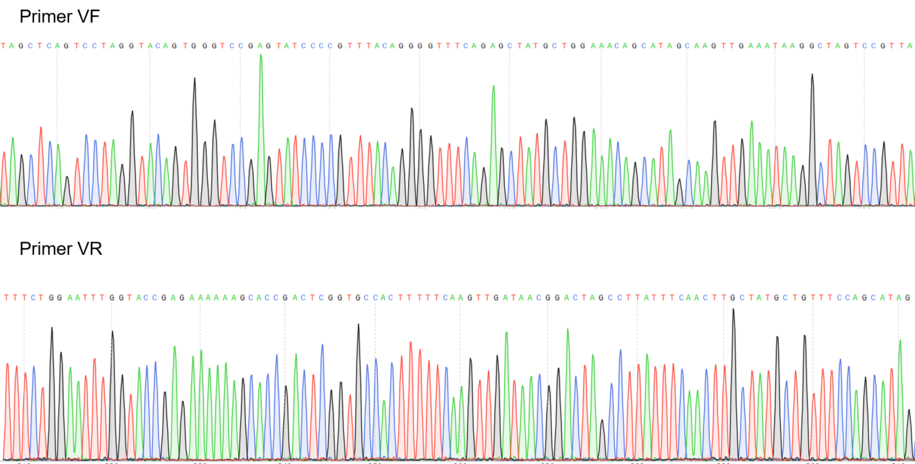 Figure 1: Sequencing electropherogram from BBa_K2457002.
Figure 1: Sequencing electropherogram from BBa_K2457002.
Figure 2: Alignment of the designed sequence and our final construction from BBa_K2457002.
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000COMPATIBLE WITH RFC[1000]
| None |