This experience page is provided so that any user may enter their experience using this part.
Please enter
how you used this part and how it worked out.
Applications of BBa_M0052
User Reviews
UNIQ9f93f744b068e321-partinfo-00000000-QINU
UNIQ9f93f744b068e321-partinfo-00000001-QINU
|
••••
iGEM14_Edinburgh
|
iGEM14_Edinburgh added this tag to RFP (Part:BBa_K1399003) and GFP (Part:BBa_K1399008) and constructed lactose/IPTG inducible measurement pathway (Part:BBa_K1399013 and Part:BBa_K1399018, respectively) to characterize and compare the instability of this SsrA-DAS tagged RFP and GFP to others containing different variants of SsrA tags. We determined the steady state fluorescence/OD ratio of E. coli JM109 at uninduced (minimum steady state) and induced (maximum steady state) state. To see how the temperature of environment can affect protein degradation efficiency we repeated experiments in several temperatures (28⁰C, 32⁰C, 37⁰C and 42⁰C). Our data are summarised below. (For more information on SsrA degradation tag system, our experimental assays and results please visit our wiki page [http://2014.igem.org/Team:Edinburgh].)
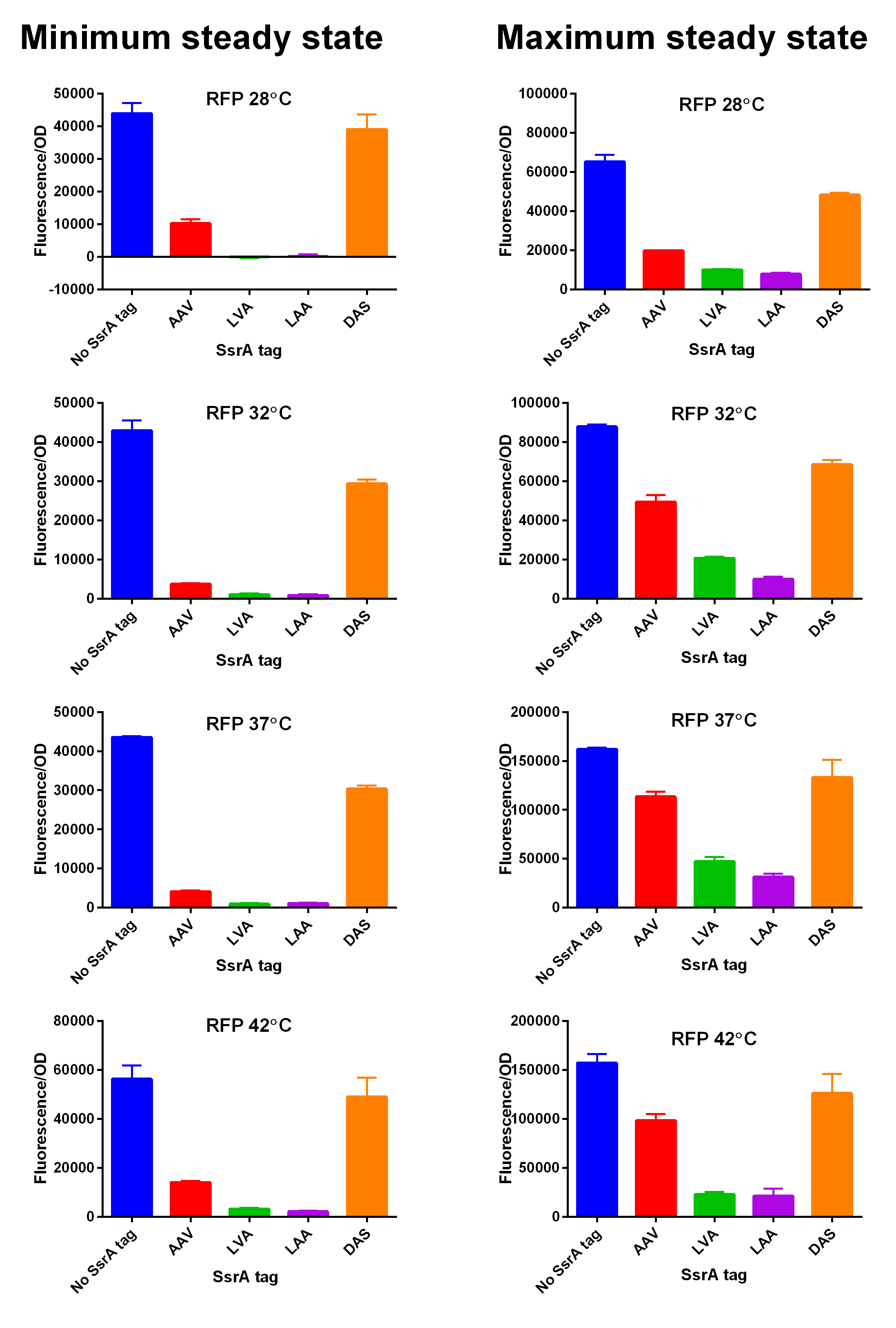 SsrA degron tagged RFP minimum (left) and maximum (right) fluorescence per OD at four different temperatures (28⁰C, 32⁰C, 37⁰C and 42⁰C).  Summary of BioBricks to which our characterization data is applicable. 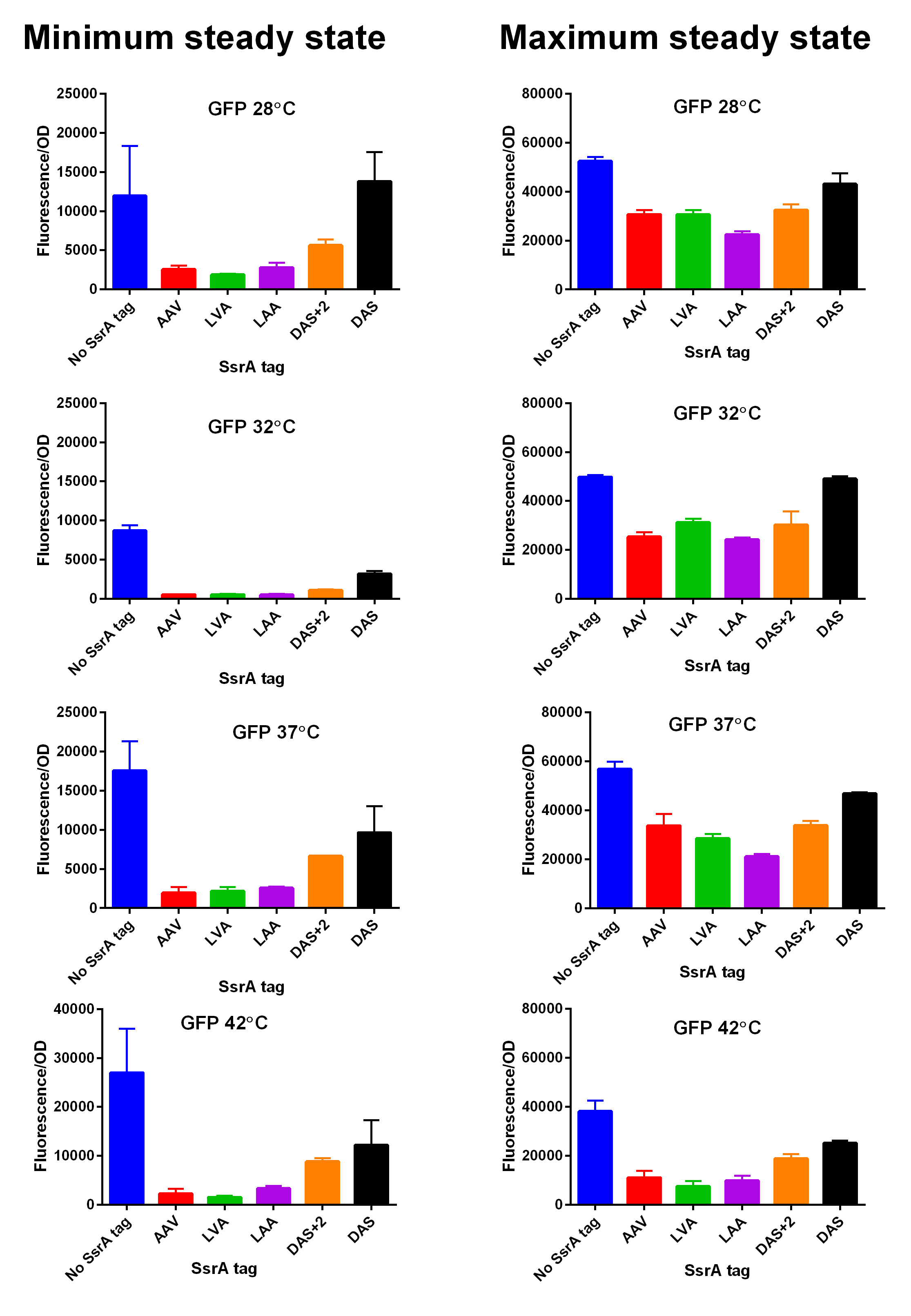 SsrA degron tagged GFP minimum (left) and maximum (right) fluorescence per OD at four different temperatures (28⁰C, 32⁰C, 37⁰C and 42⁰C).  Summary of BioBricks to which our characterization data is applicable. 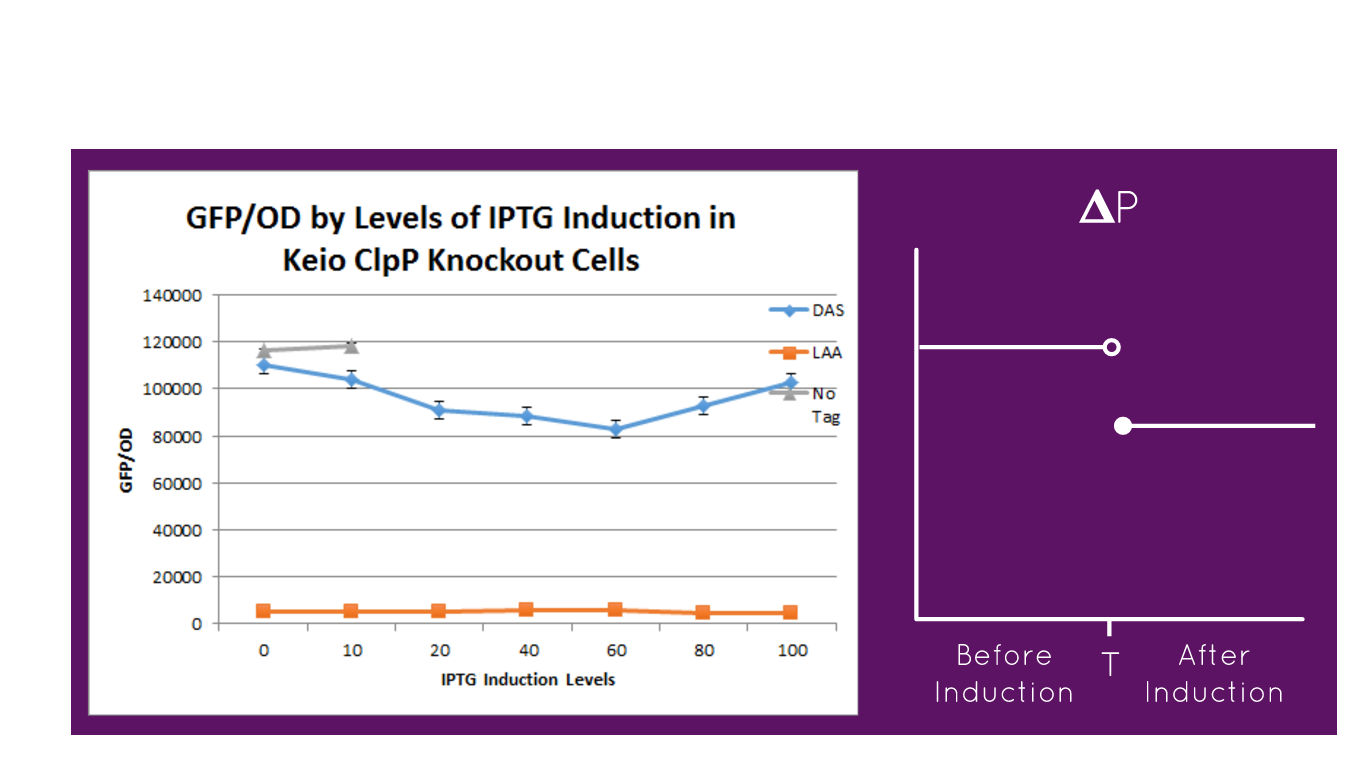
The graph depicts the experiment in which our full construct with GFP (no tag, DAS tag, LAA tag) was grown in the Keio ClpP Knockout E. coli strains. Then, it was induced with various IPTG induction levels to allow for the expression of the ClpXP degradation system BBa_K1911001 and the GFP/OD level was observed in each the E. coli cells. The expected results of the experiment were that the no tag would have no decrease in the GFP/OD level as IPTG increased, DAS tag would have a moderate decrease in the GFP/OD level as IPTG increased but not as much of a decrease relative to the Keio Wild Type E. coli cells experiment, and LAA tag would have stronger decrease in the GFP/OD level as IPTG increased but not as much of a decrease relative to the Keio Wild Type E. coli cells experiment. In the actual experiment, the results showed that the GFP/OD level for no tag relatively stayed the same when IPTG increased which was expected, the GFP/OD level for DAS tag decreased when IPTG increased which was expected, and the GFP/OD level for LAA tag decreased more relative to the DAS tag when IPTG increased which was also expected. The no tag data did not have as many data points as the LAA and DAS tag data because there was no growth for the E. coli colonies during the 30-80 micromolar IPTG levels. We theorize this is due to improper inoculation from the colonies. The Keio Wild Type inherently has the ClpXP system existing inside its E. coli cells. The graph above characterizes the BioBrick part M0052 DAS tag because by comparing the degradation of the three different constructs (no tag, DAS tag, LAA tag) we demonstrated that the relative degradation strength of the LAA and DAS tags were indeed what they were supposed to be (LAA- strong, DAS- moderate).
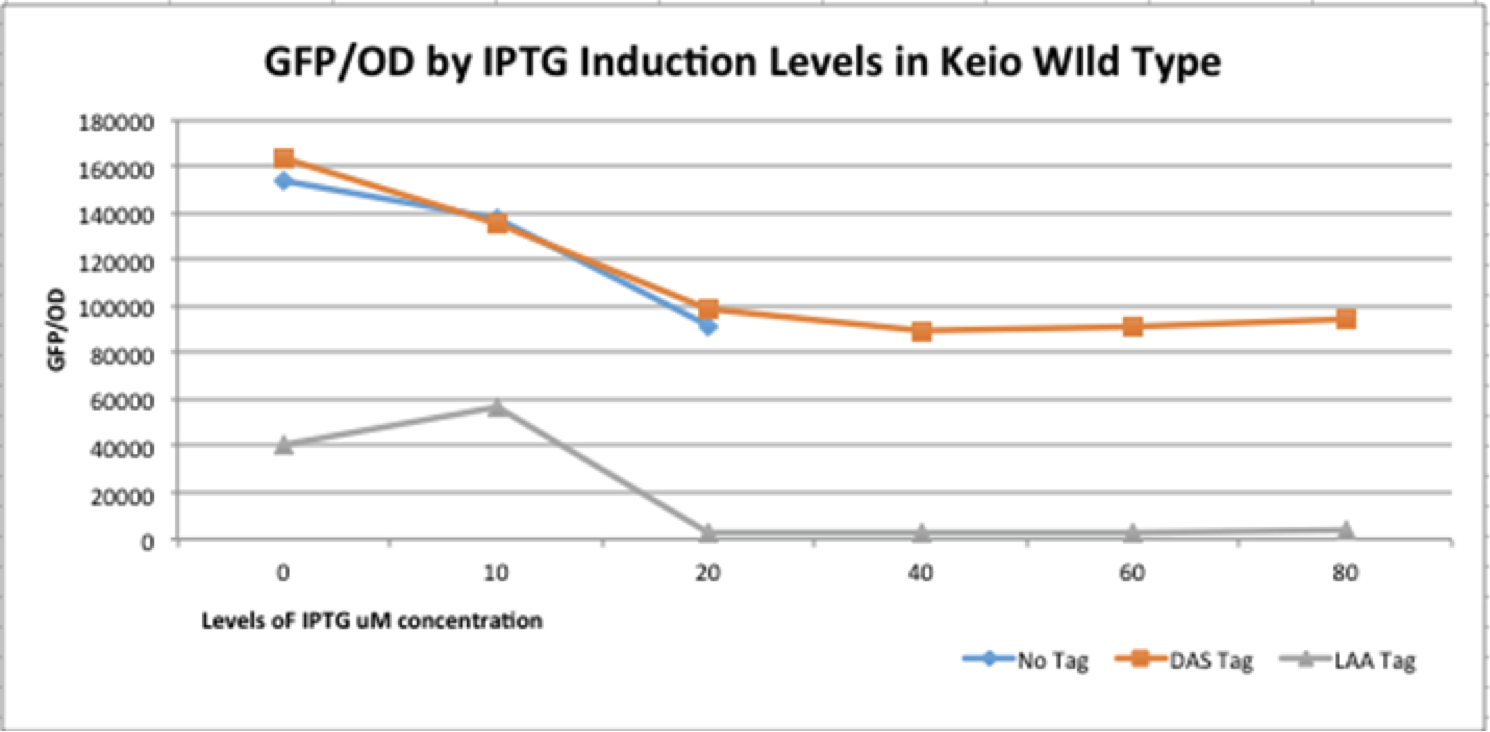
The graph depicts the experiment in which our full construct with GFP (no tag, DAS tag, LAA tag) was grown in the Keio Wild Type E. coli strains. Then, it was induced with various IPTG induction levels to allow for the expression of the ClpXP degradation system BBa_K1911001 and the GFP/OD level was observed in each the E. coli cells. The expected results of the experiment were that the no tag would have no decrease in the GFP/OD level as IPTG increased, DAS tag would have a moderate decrease in the GFP/OD level as IPTG increased, and LAA tag would have stronger decrease in the GFP/OD level as IPTG increased. In the actual experiment, the results showed that the GFP/OD level for no tag decreased when IPTG increased which was not expected, the GFP/OD level for DAS tag decreased when IPTG increased which was expected, and the GFP/OD level for LAA tag decreased more relative to the DAS tag when IPTG increased which was also expected. The no tag data did not have as many data points as the LAA and DAS tag data because there was no growth for the E. coli colonies during the 30-80 micromolar IPTG levels. We theorize this is due to improper inoculation from the colonies. The Keio Wild Type inherently has the ClpXP system existing inside its E. coli cells. The graph above characterizes the BioBrick part M0052 DAS because by comparing the degradation of the three different constructs (no tag, DAS tag, LAA tag) we demonstrated that the relative degradation strength of the LAA and DAS tags were indeed what they were supposed to be (LAA- strong, DAS- moderate) and that the ClpX subunit works in conjunction with the ClpP subunit to degrade the GFP reporter gene.
|






