Part:BBa_K1907001
Microcystinase (MlrA) for S. cerevisiae
Introduction
This part can be used to introduce microcystinase (MlrA) from Sphingomonas sp. ACM-3962 to yeast. MlrA is an enzyme that degrades some cyanobacterial toxins, most notably microcystin-LR (MC-LR). To facilitate detection and purification of the enzyme, a 6X Histidine tag is fused on the C-terminus of MlrA, with a short linker in between. The full protein-coding sequence has been codon-optimized for expression in Saccharomyces cerevisiae by GeneArt.
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21INCOMPATIBLE WITH RFC[21]Illegal BglII site found at 166
Illegal BglII site found at 907 - 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000INCOMPATIBLE WITH RFC[1000]Illegal BsaI.rc site found at 849
Background
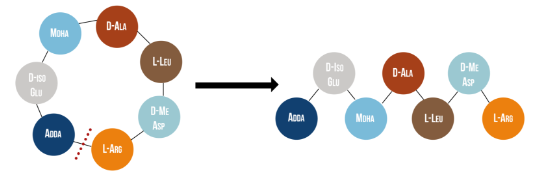
Microcystinase, or MlrA, is a member of a set of enzymes that degrade microcystins. These enzymes - MlrA, MlrB, MlrC and MlrD - are found naturally in some gram-negative bacteria, for example Sphingomonas sp. They each have a different function in the overall process of microcystin degradation. MlrA is first in the sequence to act. It hydrolyzes the peptide bond between the Adda and arginine amino acids in microcystin-LR (MC-LR). The breakage of this bond linearizes the toxin (Figure 1). Following this, MlrB (a serine protease) and MlrC (a metalloprotease) are able to further degrade the linearized product into individual amino acids, which are then used in the cell’s metabolism. Therefore, the toxin is likely used as a carbon source. MlrD’s function is not fully understood at this point, but it may have a role in importing the toxins. (Bourne et al., 2001)
Based on a homologous structure model (Figure 2) obtained from the Phyre2 tool (Kelley et al., 2015), it seems that the enzyme is a transmembrane protein consisting of eight hydrophobic α-helices that form a channel, with the presumed active site (Bourne et al., 2001) residing in the middle. It is worth pointing out, however, that the model does not cover the whole length of the MlrA sequence, as only very few homologs were found for the sequence - the homology model is mostly based on two homologs that didn’t cover the very ends of either terminus. Other than this evidence, there are previous studies pointing to the direction of MlrA being a membrane protein (Pei et al., 2011). However, when the enzyme was heterologously produced in E. coli , it was noticed that it had localised mostly in the soluble fraction of the cytosol, in an active state (Dziga et al., 2012).
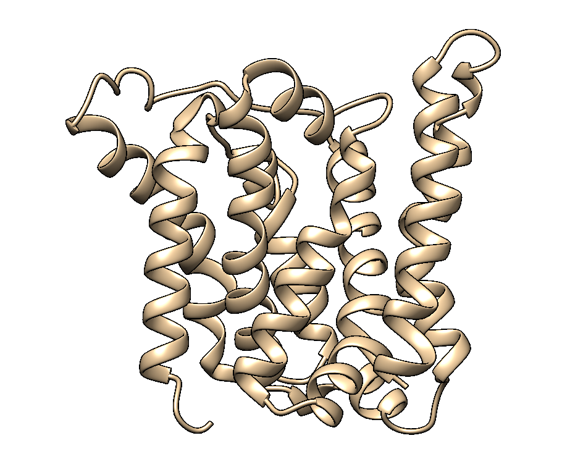
The mechanism of MlrA’s function is not yet fully understood. The homologous structure model gives some suggestions as to how the enzyme might work: it seems that one end of the channel is wider than the other, and that the active site resides in the middle of it. Consequently, if the enzyme is located on the outer membrane of the cells, it might actually be a transporter which the cells use to import the toxin and simultaneously linearize it.
Previous use
This part was used in S. cerevisiae strain SS328-leu. It was used in the pRS415-GAL plasmid backbone, with the insert being cloned into the plasmid using the SpeI and XhoI restriction sites (5' and 3', respectively). These restriction sites flanked the protein-coding site directly. The part was modified to be flanked by the BioBrick standard prefix and suffix for shipping to the registry.
Expression
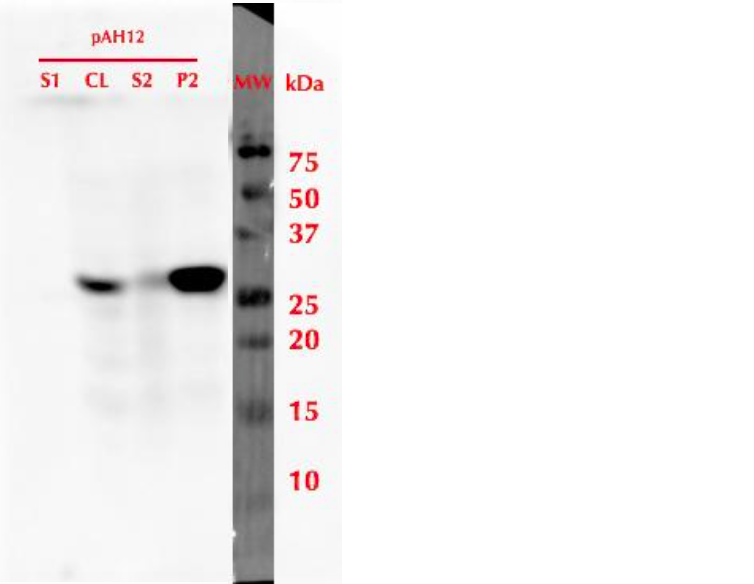
To produce the protein, a 20 mL pre-culture was prepared in SD-leu + 2% glucose and grown overnight at 30 °C. The culture was then diluted into a final volume of 500 mL using SD-leu + 0.5% glucose, to an OD600 value of 0.2. This culture was refreshed in 30 °C for 6 h, and then induced by adding galactose to a final concentration of 2%. Expression was continued overnight, then cells were harvested. Samples from different stages of cell culture and cell lysis were collected for SDS-PAGE and analysis with western blot utlizing anti-His antibodies. The results of this analysis are presented in figure 3. It was observed that after cell lysis, the protein localized mainly in the insoluble fraction.
Localization
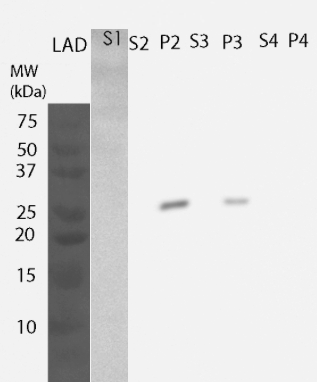
For determining the location of the protein, yeast cells were lysed and fractions were further processed to obtain information on the different cellular components. All of these fractions were then analyzed by western blotting (Figure 4). MlrA was detected in the fraction comprising inclusion bodies, plasma membrane and cell wall (P2 fraction) as well as fraction containing only cell wall and plasma membrane (P3 fraction), indicating that the protein is localized in the plasma membrane. Unlike previously, no enzyme was detected in the soluble fraction.
Activity measurements
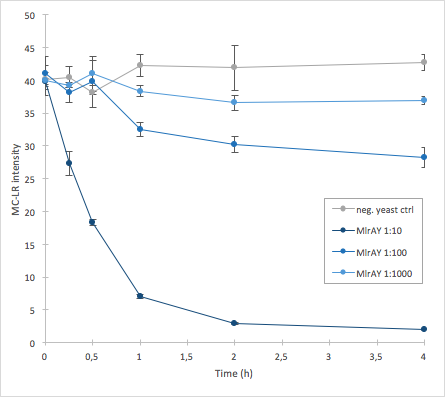
The activity of MlrA was measured by incubating the enzyme in different dilutions with microcystin extract. The final concentration of microcystin-LR was estimated to be 1 μg/L. Samples were taken at different time points and they were analysed with liquid chromatography/mass spectrometry (LC-MS). This gives data of the relative abundance of microcystin-LR in each sample. The results were then plotted against time, together with a negative control which consisted of microcystin extract and cell lysate from S. cerevisiae SS328-leu strain which did not have an MlrA construct.
The results we acquired from this construct show that the amount of microcystin decreases, which means that the enzyme is active (Figure 5). The results also show that for example the 1:10 dilution degrades the microcystin-LR about ten times faster than the 1:100 dilution.
For more details on the experiments done with this BioBrick, visit <a href = "http://2016.igem.org/Team:Aalto-Helsinki/Laboratory" target="blank" > http://2016.igem.org/Team:Aalto-Helsinki/Laboratory</a>
References
Bourne DG, Jones GJ, Blakeley RL, Jones A, Negri AP, Riddles P., 1996. Enzymatic Pathway for the Bacterial Degradation of the Cyanobacterial Cyclic Peptide Toxin Microcystin LR. Appl. Environ. Microbiol. 62(11), pp. 4086-4094.
Bourne DG, Riddles P, Jones GJ, Smith W, Blakely RL., 2001. Characterisation of a gene cluster involved in bacterial degradation of the cyanobacterial toxin microcystin LR. Environ Toxicol. 16(6), pp. 523-34.
Dziga D, Wladyka B, Zielińska G, Meriluoto J, Wasylewski M., 2012. Heterologous expression and characterisation of microcystinase. Toxicon, 59(5), pp. 578-586.
Kelley LA, Mezulis S, Yates CM, Wass MN, Sternberg MJE., 2015. The Phyre2 web portal for protein modeling, prediction and analysis. Nature Protocols 10, pp. 845–858.
Pei J, Mitchell DA, Dixon JE, Grishin NV., 2011. Expansion of type II CAAX proteases reveals evolutionary origin of g-secretase subunit APH-1. J. Mol. Biol. 410:18–26.
//chassis/eukaryote/yeast
| None |