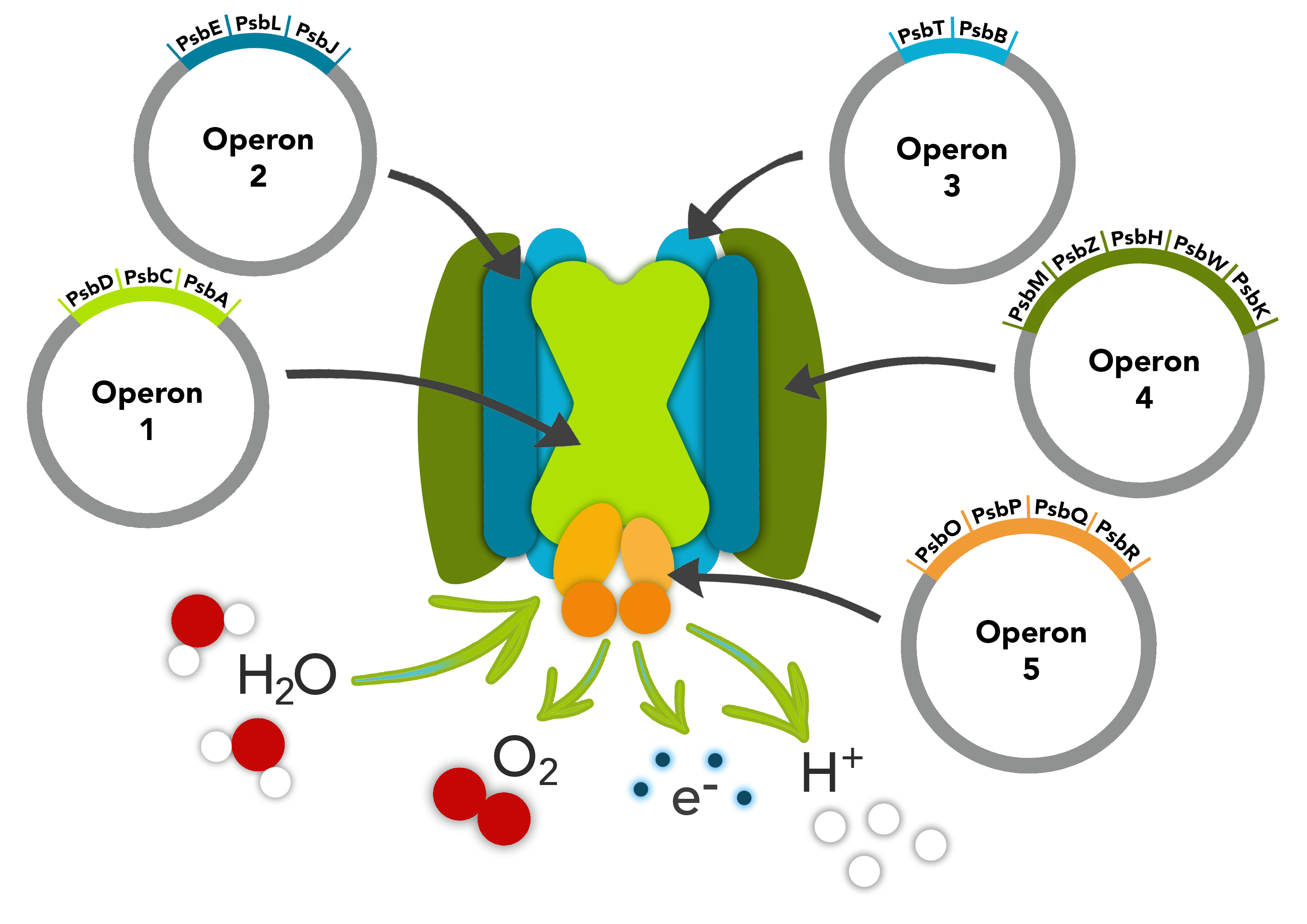
Part:BBa_K1998006
psbOPQR
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21INCOMPATIBLE WITH RFC[21]Illegal BglII site found at 1998
Illegal BglII site found at 2327 - 23COMPATIBLE WITH RFC[23]
- 25INCOMPATIBLE WITH RFC[25]Illegal NgoMIV site found at 754
Illegal NgoMIV site found at 1400 - 1000INCOMPATIBLE WITH RFC[1000]Illegal BsaI site found at 2391
Overview
This part is composed of the psbO, psbP, psbQ and psbR genes. The psbO gene that comprises this part ecodes a manganese stabilizing protein. The psbP gene that comprises this part transcribes a protein that optimizes the availability of Ca2+ and Cl- cofactors in the Oxygen Evolving Complex in PSII to maintain the active Manganese cluster. The psbQ protein is an oxygen enhancer protein. The psbR protein is an important link in the Photosystem II core complex for stable assembly of the Oxygen Evolving Complex.
These parts make up one of the operons in our PSII pathway.
Biology & Literature
PsbO encodes psbO, a 33 kDa protein which acts to stabilise the cluster of four Mn2+ which forms the catalytic centre of the oxygen evolving complex (OEC) (Murata & Miyao, 1985). PsbO consists of an eight strand β-barrel, with a large loop between strands five and 6, which connects the OEC to the luminal surface (Ferreira, Iverson, Maghlaoui, Barber, & Iwata, 2004). Deletion studies of psbO in Synechocystis sp. have shown little effect to photoautotrophic growth and oxygen evolution, however a markedly higher susceptibility to photoinhibition (Mayes, Cook, Self, Zhang, & Barber, 1991). This suggests PsbO is not essential to PSII assembly or water-splitting, however provides protection of PSII from light-induced damage.
The psbP gene that comprises this part transcribes a protein that optimizes the availability of Ca2+ and Cl- cofactors in the Oxygen Evolving Complex in PSII to maintain the active Manganese cluster.
The psbQ gene encodes a 17 kDa protein that is located on the inner side of the thylakoid lumen. Its N-terminus region contains a polyproline II helix (Balsera, M., et al., 2005). It plays an important role in the ability of photosystem II to function in low light conditions (Yi, X., et al., 2006). Studies involving mutant cells have shown that, in the absence of psbQ, their ability to generate oxygen is impaired and psbV is destabilised (Kashino, Y., et al., 2006).
The psbR gene encodes a 10 kDA protein that is essential to the stable assembly of psbP, which is a component of the oxygen-evolving complex of photosystem II. The assembly of psbR is mediated by psbJ (Suorsa, M., et al., 2006). Electron transport in both the donor and the acceptor side of the photosystem II core complex are modified in the absence of this protein (Allahverdiyeva, Y., et al., 2007).
Protein information
psbO
mass: 27.96kDa
sequence:
MAQKVGQAAAAAALATAMVAGSANALTFDEIQGLTYLQVKGSGIANTCPVLESGTTNLKELKAGSYKLENFC IEPTSFTVKEESQFKGGETEFVKTKLMTRLTYTLDAMSGSFKVGSDGSAELKEDDGIDYAATTVQLPGGERV AFLFTIKQFDGKGTLDNIKGDFLVPSYRGSSFLDPKGRGGSTGYDNAVALPARADAEELLKENVKITKALKG
SAVFSVAKVDPVTGEIAGVFESIQPSDTDLGAKPPKDIKVTGLWYAQLK*
psbP
mass: 23.17kDa
sequence:
MASGSDVSRRAALAGFAGAAALVSSSPANAAYGDSANVFGKVTNKSGFVPYAGDGFALLLPAKWNPSKENDF PGVILRYEDNFDAVNNLVVIAQDTDKKAIADFGSQDKFLESVSYLLGKQAYSGETQSEGGFAPNRVSAASLL DVSTTTDKKGKTYYKYELLVRSADGDEGGRHQLIGATVGSDNKLYIIKIQIGDKRWFKGAKKEAMGAFDSFTVV*
psbQ
mass: 19.6kDa
sequence:
MASGESRRAVLGGLLASAVAAVAPKAALALTPVDLFDDRSVRDRGFDLIYEARDLDLPQNVREGFTQARASL DETKKRVKESEARIDADLDVFIQKSYWTEAREQLRRQVGTLRFDLNTLASTKEKEAKKAALGLRKEFIQAVED
LDFALREKDQASAAKKLEITKAKLDSVLAAVL
psbR
mass: 12.2kDa
sequence:
MGGGKTDITKVGLNSIEDPVVKQNLMGKSRFMNKKDWKDASGRKGKGYGVYRYEDKYGANVDGYSPIYTPDL
WTESGDSYTLGTKGLIAWAGLVLVLLAVGVNLIISTSQLGA
References
| None |
