Part:BBa_I721002
Lead Binding Protein
lead binding protein part 3
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000COMPATIBLE WITH RFC[1000]
Part BBa_1721002 is the Lead Binding Protein
USE AND BIOLOGY
iGEM16_HSiTAIWAN: Measure PbrR(I721002) generation through GFP expression: (1) PbrR under autoregulation of Ptet (BBa_K1961004) and (2) PbrR under control of TetR, which is under control of a constitutive promoter (BBa_K1961005)

Figure 1. The Constitutive circuit and the related rate equations

Figure 2. The Autoregulation circuit and the related rate equations

Figure 3. The parameter space for constitutive circuit

Figure 4. The fold analysis for autoregulation and constitutive circuits
Description
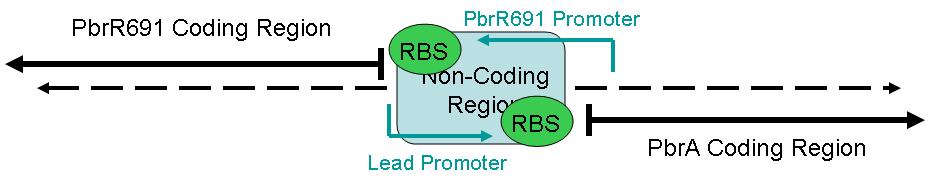
This part is designed to enable Lead Detection in a cell. When Lead (Pb +2) enters the cell, it will couple with the Lead Binding protein, forming a dimer. BBa_I721002 contains only the Lead Binding Protein sequence, and must be combined with a Lead Promoter sequence in ordered to be remotely functional.
This sequence was obtained using PCR from the genome of Ralstonia metallidurans. Biobrick cut sites were added using specific primers during PCR.
This part is not known to function. "PbrR691/promoter combinations in presence of lead nitrate give no GFP production and no AHL production compared to control. "
The figure below illustrates the layout of this sequence.
BBa_I721002 contains only the Lead Binding Protein region.
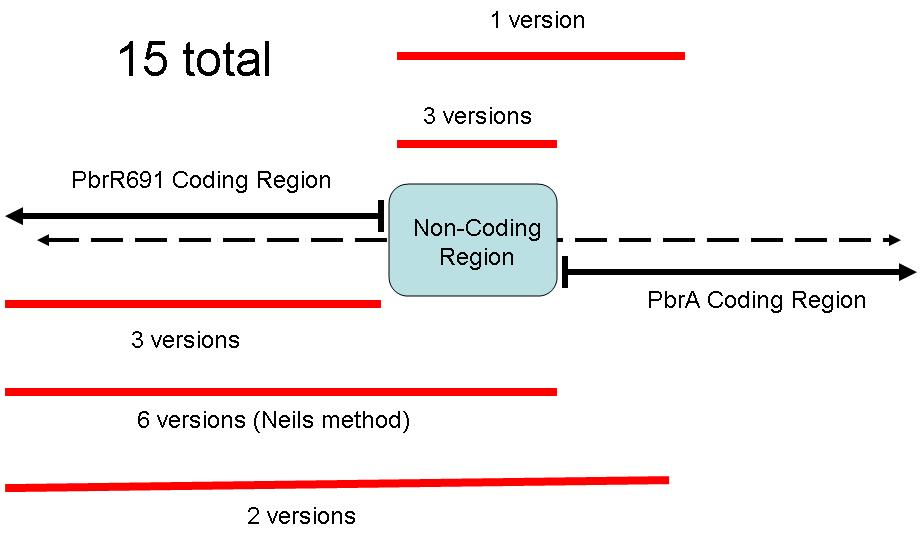
Because of the different coding regions contained in this sequence, the Brown iGEM Team chose 15 different regions of the Pbr691 and PbrR691 sequences to extract. We attempted to create Biobricks from each of these 15 different sequences. Some contained only the PbrR Lead Binding Protein, some contained only the Pbr Promoter, and some parts contained both. The following diagram illustrates these different parts.
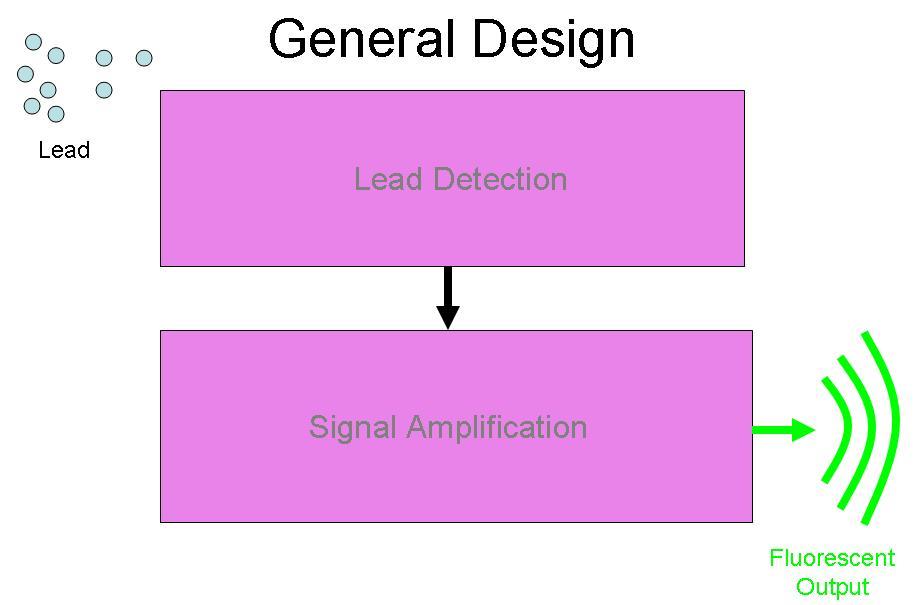
BBa_I721002 was designed to be a part of the Brown University Cellular Lead detector device. The BBa_I721002 Lead Binding Protein was designed to be attached with part BBa_I721004, the Lead Promoter. This was designed to form a Lead Detection System, to be attached to the Amplification system by Imperial College, (BBa_J37015), with GFP as the output of the device. The schematic for this project is below:
//cds/receptor
| None |

 1 Registry Star
1 Registry Star