Part:BBa_K1639005
H-NS(T108I)
global DNA-binding transcriptional dual regulator H-NS mutant form.
Usage and Biology
Donato and Kawula investigated the effect on bacterial motility of random point mutations in the HNS protein. As a result of their Swarm plate assay and experiments for the determination of flagellar rotational speed, it was found that some random mutations (HSN-T108I and HNS-A18E) resulted in bacteria with a stronger and faster flagella as compared with wild type bacteria. This single amino acid change caused a 50% increase in the binding of HNS to FliG.. It was also shown that this single amino acid change in HNS resulted in an approximately 50% increase in flagellar rotation speed and about a 2-fold increase in the bacteria’s swarm size.



In the results from fluorescence anisotropy and chemical cross-linking, Donato and Kawula showed that there is an interaction between HNS and FliG, the protein responsible for flagellar torque; and they show that the mutant form of HNS binds FliG 50% more often than does wild-type HNS: They explain the increase in the bacteria’s swarm rate and swim speed caused by mutant HNS thusly “We position H-NS at the interface between the rotor and stator, directly linked to the C terminus of FliG(Fig. 5 A). Tighter binding of mutant H-NST108I toFliG (Fig.5 B) may cause increases in flagellar speed by altering the conformation of FliG relative to the other rotor proteins and/or the MotA·B complex, thus, compacting the motor complex and allowing fast errotation by creating less friction within the surrounding stationary MotA·B ring complex (56).
This study having captured our attention, we decided to use mutant HNS to increase flagellar speed and torque power.
Characterization
SOFT AGAR PLUG ASSAY
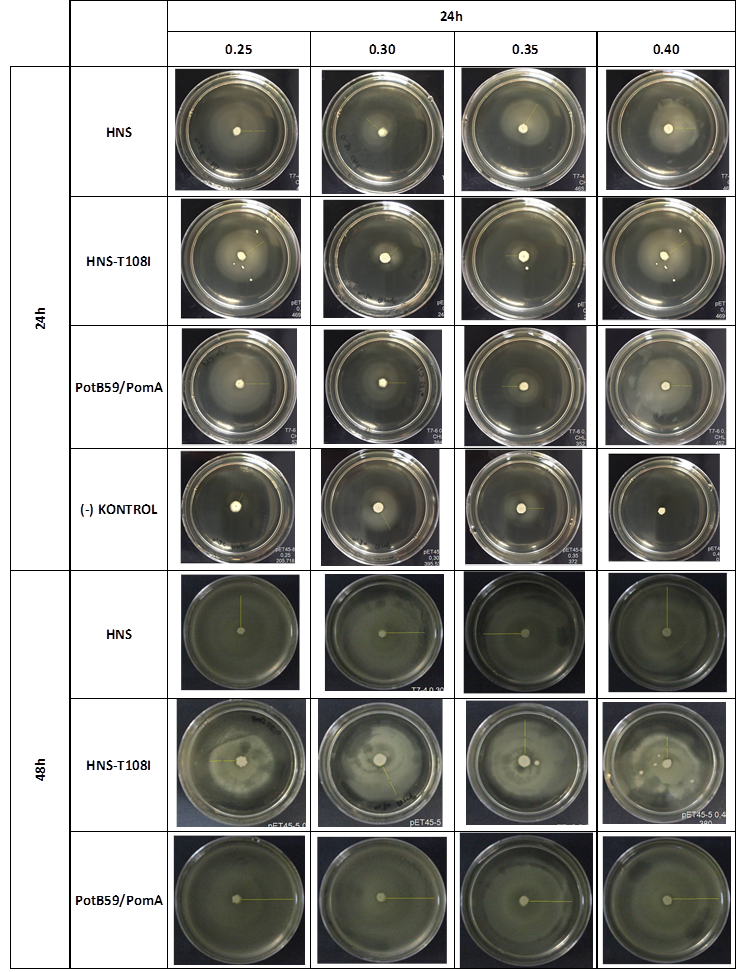
After showing that we produced required proteins successfully, in order to learn if they are functional, we did a motility assay called ‘plug assay’. We inoculated bacteria in different concentrations into the soft agars and incubated them at 37°C. After this, we measured their colony diameters at 24. and 48. hours. The results are shown below.
plasmid transformed bacteria HNS:pET45-b-HNS plasmid transformed bacteria HNS-T108I:pET45-b-HNS-T108I plasmid transformed bacteria PotB59/PomA :pET45-b-PotB59/PomA plasmid transformed bacteria.The measurements of 0.25 and 0.4 concentrations in first 24. hour period, gives us significant positive results . If we compare the results,colony diameters are ordered like this; (-) kontrol < Wild type HNS < PotB59/PomA. The results of 0.30 and 0.35 agar concentrations doesn’t give significant information. Besides, in 48. hour of incubation the diameter of colonies reached to plate’s diameter, so measurement results didn’t show any difference than 24. hour’s.
FUNCTIONAL ASSAY
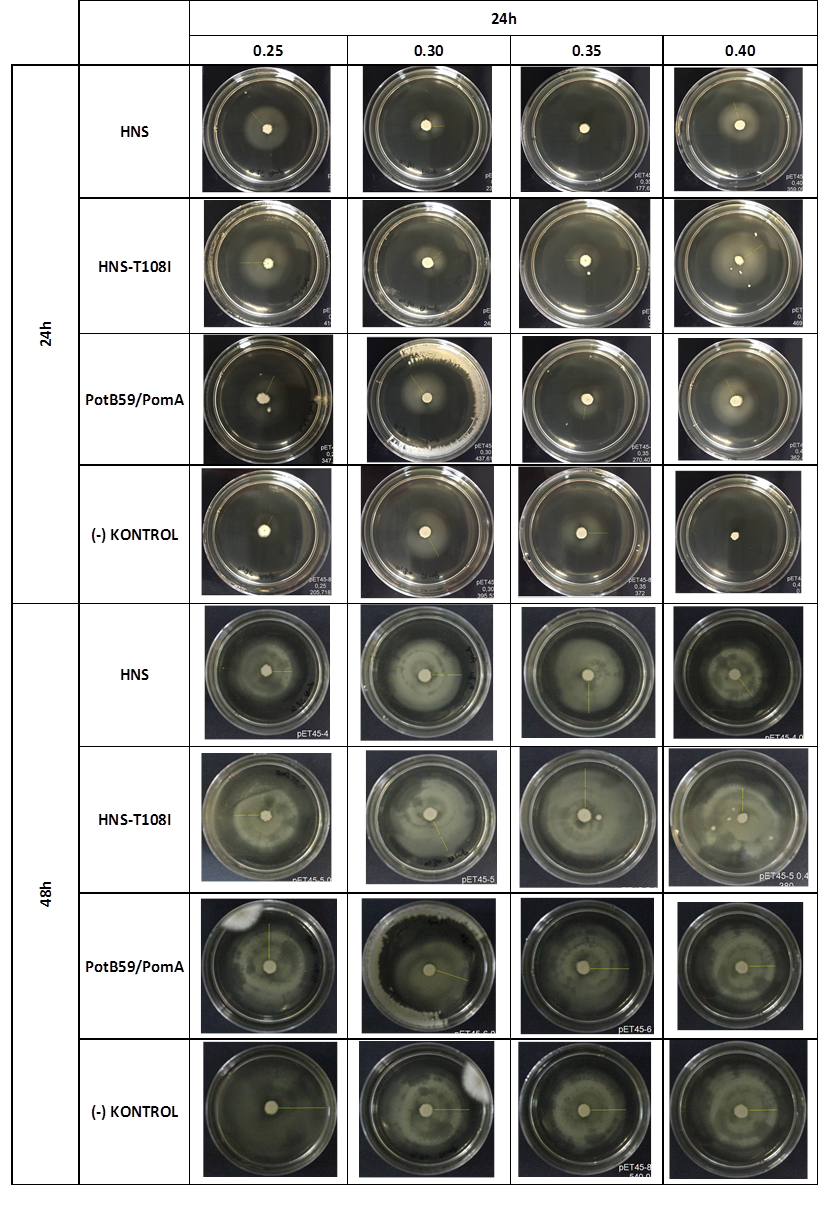
We repeated functional assays after we cloned the genes into PSB1C3-T7. The results are shown below.
The measurements which made at first 24. hour period, gives us significant positive results . If we compare the results,colony diameters are ordered like this; (-) control < Wild type HNS < HNS-T108I< PotB59/PomA
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21INCOMPATIBLE WITH RFC[21]Illegal BamHI site found at 2
Illegal XhoI site found at 426 - 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000INCOMPATIBLE WITH RFC[1000]Illegal SapI.rc site found at 162
References
[1]Gina M. Donatoand Thomas H. Kawula(1998)TheJournalBiologicalChemistryVol. 273, No. 37, 24030-24036 [2]Macnab, R. M. (1992) Annu. Rev. Genet. 26, 131–158 [3]Macnab, R. M. (1996) EscherichiacoliandSalmonellatyphimurium Cellular andMolecularBiology (Neidhardt, F. C.,Curtiss, R., III, Ingraham, J. L., Lin, E. C. C.,Low, K. B., Magasanik, B., Reznikoff, W. S., Riley, M., Schaechter, M.,andUmbarger, H. E., eds) 2nd Ed., pp. 123–145, American SocietyforMicrobiology, Washington, D. C. [4]Blair, D. F.,andBerg, H. C. (1990) Cell 60, 439–449 [5]Blair, D. F.,andBerg, H. C. (1991) J. Mol. Biol. 221, 1433–1442 [6]Garza, A. G., Harris-Haller, L. W., Stoebner, R. A., andManson, M. D. (1995) Proc. Natl. Acad. Sci. U. S. A. 92, 1970–1974 [7]Marykwas, D. L.,Schmidt, S. A., andBerg, H. C. (1996) J. Mol. Biol. 256,564–576 [8]Tang, H.,Braun, T. F., andBlair, D. F. (1996) J. Mol. Biol. 261, 209–221 [9]Yamaguchi, S.,Fujita, H., Ishihara, A., Aizawa, S.-I., andMacnab, R. M. (1986) J. Bacteriol. 166, 187–193 [10]Irikura, V. M.,Kihara, M., Yamaguchi, S., Sockett, H., andMacnab, R. M. (1993) J. Bacteriol. 175, 802–810 [11]Lloyd, S. A.,Tang, H., Wang, X., Billings, S., andBlair, D. F. (1996) J. Bacteriol. 178, 223–231 [12]Welch, M.,Oosawa, K., Aizawa, S.-I., andEisenbach, M. (1993) Proc. Natl. Acad. Sci. U. S. A. 90, 8787–8791 [13]Vogler, A. P.,Homma, M., Irikura, V. M., andMacnab, R. M. (1991) J. Bacteriol. 173, 3564–3572 [14]Tang, H.,Billings, S., Wang, X., Sharp, L., andBlair, D. F. (1995) J. Bacteriol. 177, 3496–3503 [15]Asai, Y., T. Yakushi, I. Kawagishi, and M. Homma. 2003. Ion-couplingdeterminants of Na + -drivenand H + -drivenflagellarmotors. J. Mol. Biol. 327: 453-
| n/a | H-NS(T108I) |