Part:pSB1C3
Coding sequence of Nluc398-FRB
Nluc 398 fusion protein ORF
Firefly (Photinus pyralis) luciferase can be split to N terminal (Nluc) and C terminal (Cluc) fragments and each of them is inactive. When they two reassembled non-covalently, the enzymatic activity would be reconstituted and the recovered luciferase is able to oxidize luciferin and produce detectable bioluminescence. Currently there are different combinations of split fragments, among which Nluc 416 / Cluc 398 and Nluc 398/ Cluc 394 are widely used[1].
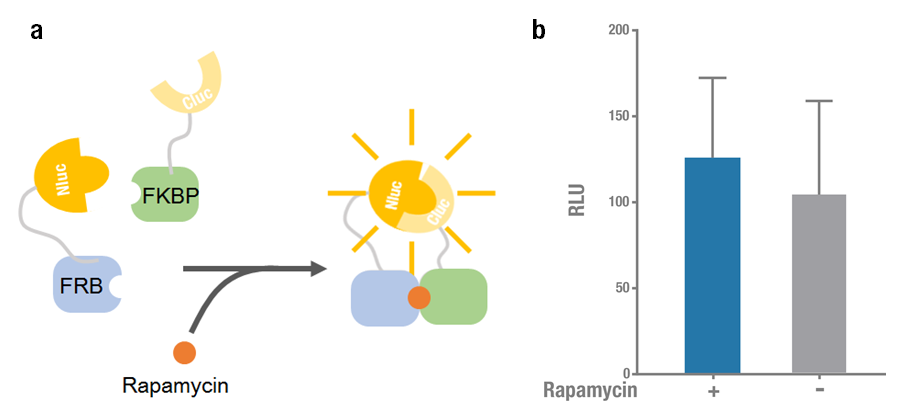
Rapamycin-binding domain (FRB) of human mTOR (mammalian Target of Rapamycin) binds with high affinity to FK-506-binding protein 12 (FKBP). Rapamycin is able to induce the dimerization to form a FRB-rapamycin-FKBP complex[2]. This protein-protein interaction can be visualized by split luciferase[3]. FRB and FKBP are fused to Nluc and Cluc respectively, and adding rapamycin can induce the approaching and reconstitution of split luciferase (Figure 1a).
2015 Peking iGEM fused Nluc 398 to N terminus of FRB (Nluc 398-FRB, BBa_K1689004) and combined it with FKBP-Cluc 394 (BBa_K1689006) to validate the functional reconstitution of split luciferase. However, compared with Nluc 416/ Cluc 398, the bioluminescence intensity didn't increase significantly after rapamycin was added (Figure 1). Therefore we discarded them and chose Nluc 416/Cluc 398 as our split luciferase in the project (See BBa_K1689003 or BBa_K1689005).
Figure 1. Rapamycin-induced N-luc-FRB/FKBP-C-luc complementation. (a) The working mechanism of rapamycin induced dimerization. The interacting protein partners (FRB & FKBP) get closer and dimerize soon after rapamycin is added (40nM) [4], thus to reconstitute the enzymatic activity of luciferase. (b) The experimental data. Error bars denote s.d.; n=3.
References
1. Ramasamy Paulmurugan, Sanjiv S. Gambhir. Firefly Luciferase Enzyme Fragment Complementation for Imaging in Cells and Living Animals. Anal Chem. 2005 March 1; 77(5): 1295–1302.
2. Rivera, V. M., T. Clackson, S. Natesan et al. A humanized system for pharmacologic control of gene expression. Nat. Med. 1996. 2:1028–1032.
3. Ramasamy Paulmurugan, Sanjiv S. Gambhir. Combinatorial Library Screening for Developing an Improved Split-Firefly Luciferase Fragment-Assisted Complementation System for Studying Protein-Protein Interactions. Anal. Chem. 2007, 79, 2346-2353.
4. Ramasamy Paulmurugan and Sanjiv S. Gambhir. Combinatorial Library Screening for Developing an Improved Split-Firefly Luciferase Fragment-Assisted Complementation System for Studying Protein-Protein Interactions. Anal. Chem. 2007, 79: 2346-2353.
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12INCOMPATIBLE WITH RFC[12]Illegal NheI site found at 7
Illegal NheI site found at 30 - 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000COMPATIBLE WITH RFC[1000]
//plasmidbackbone/copynumber/high
| chassis | |
| copies | 100-300 |
| insert | None |
| mcs | BioBrick |
| origin | pMB1 |
| resistance | C |

 1 Registry Star
1 Registry Star