Part:BBa_K1471003
RBS with MerB.
RBS
The initiation of protein biosynthesis is a major determinant of the efficiency of gene expression at the translational level. It is known that the nucleotide sequences around the AUG translation initiation codon act as an important signal to trigger the initiation of the translation event. (Kozak, 1987) Understanding regulatory mechanisms of protein synthesis in eukaryotes is essential for the accurate annotation of genome sequences. Kozak reported that the nucleotide sequence GCCGCC(A/G)CCAUGG (AUG is the initiation codon) was frequently observed in vertebrate genes and that this 'consensus' sequence enhanced translation initiation. However, later studies using invertebrate, fungal and plant genes reported different 'consensus' sequences. (Nakagawa, 2007)
Although for any protein analysis it is crucial to know exactly which region of the mRNA is coding for protein, prediction of the translation initial site is still an unsolved problem. In eukaryotes, the scanning model postulates that the ribosome attaches first to the 5' end of the mRNA and scans along the 5'-3' direction until it encounters the first AUG. While translation initiation from the first AUG holds true in many cases, there are also a considerable number of exceptions. In these exceptions the main determining factor in AUG choice is the context of the respective codon. (Rangan, 2008)
Two decades ago, a consensus sequence for the context of the AUG codon in higher plants was proposed on basis of very limited number of sequences. Joshi and colleagues got the generally assumption that the consensus sequence found (aaaaacaA(A/C)aAUGG) is valid for all plant clades, but Rangan found out that a considerable degree of variation between plants and major between the major eukaryotic groups along with some conserved features. However, the large variability and the periodicity suggest that general structural features rather than precise nucleotide sequence may play an important role in transcription initial site. (Rangan, 2008)
MerB
MerB belongs to the mercury resistant bacteria operon, is considered a key enzyme for the bioremediation and detoxification of various mercury compounds; being methylmercury the most notable and relevant. This gene codes for organomercury lyase, catalyzes the protonolysis of the mercury and carbon bond and release less toxic mercury specie (Hg2+).
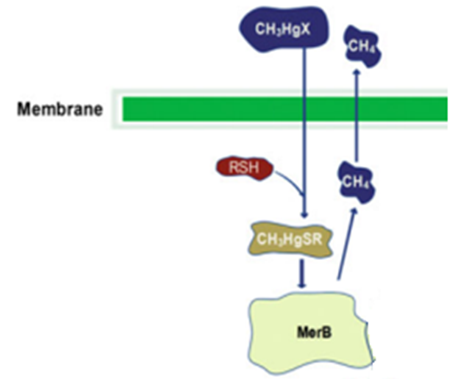
The enzyme has two conserved cysteines residue, Cys-96 and Cys-159, which behave as a substrate binding region. This region has an important role in the cleavage of the carbon-mercury bond. The Asp-99 residue of merB plays an active function in the transference of the proton during the protonolysis. Cys-117 plays the most important structural role. Other mechanisms have been proposed for the MerB function. One is the mechanism I, this declares that the methylmercury binds first to one of the two cysteines residues, the other cysteine will donate the proton of the leaving group (CH3) in the Hg-C bond cleavage. The mechanism II also refers that the methylmercury binds to one of the cysteines, however the other cysteine transfer the proton to the Asp99. This step allows to the cysteines to coordinate with the methylmercury, then the Asp99 protonates the CH3 and yields the Hg-C cleavage products.
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25INCOMPATIBLE WITH RFC[25]Illegal NgoMIV site found at 428
Illegal AgeI site found at 375 - 1000COMPATIBLE WITH RFC[1000]
| protein | |
| rbs |
