Part:BBa_K1383001
BBa_K1383001 (mCherry- Theoretical strongest RBS)
Usage and Biology
The ribosome binding sites (RBS) of archaea are not well characterized. By creating and characterizing a library of RBS sequences, researchers will be able to express proteins of interest at variable levels of expression in Methanococcus maripaludis. Ribosome binding sites are typically 6-7 base pair sequences on a transcript that is complementary to the 3’ end of the 16S rRNA. After binding of the RBS to the ribosome, translation will be initiated. An RBS with higher affinity for the ribosome will result in higher rate of translation, and inversely, an RBS with lower affinity will result in lower rate of translation.
Characterization
Characterization of the RBS sequences was accomplished using the red fluorescent protein, mCherry as a reporter. Qualitative analysis was evaluated by visualization of the RFP and quantitative analysis was completed through use of a plate reader for reading fluorescence.
WAT
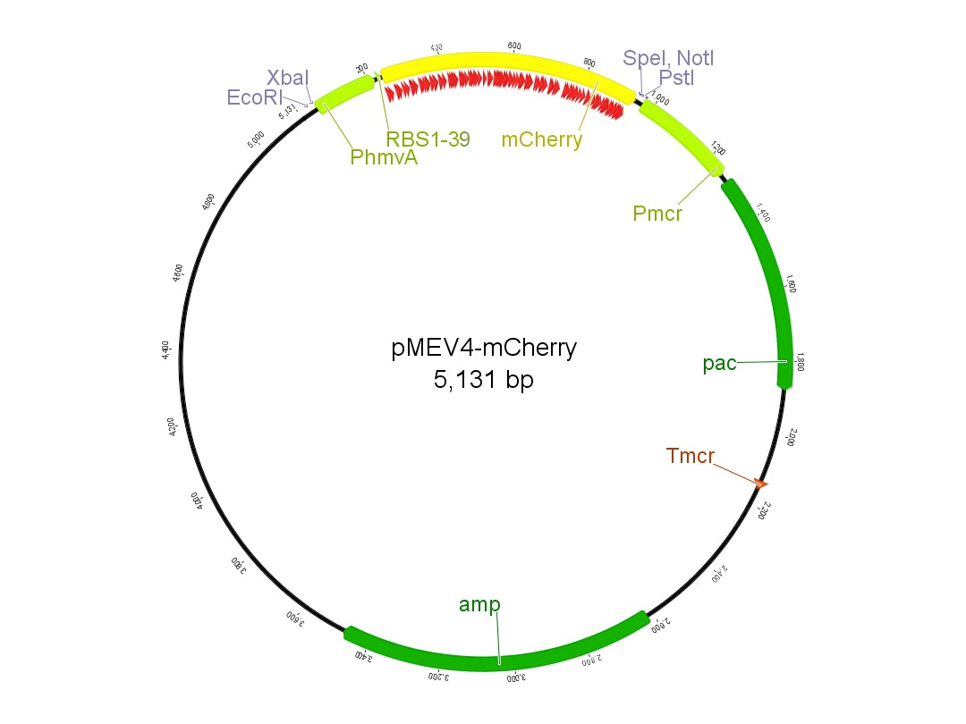
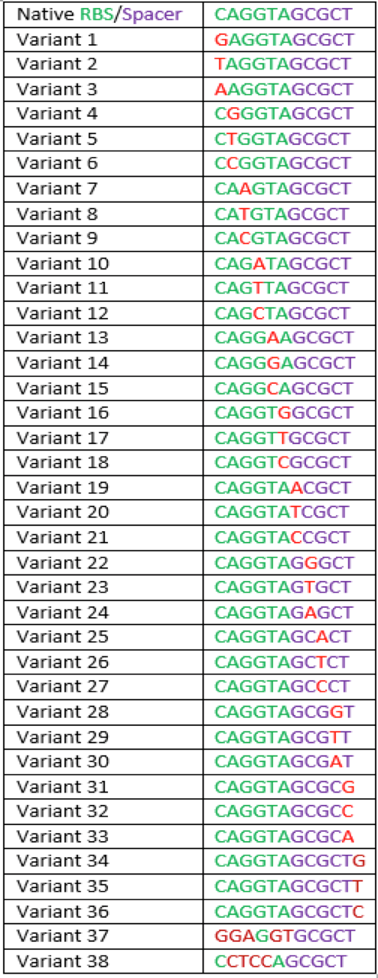
The region labeled 'RBS 1-39' in the pMEV4-mCherry vector (figure 1) is the site immediately upstream of the mCherry gene that will be subject to variation. Specifically, we will be mutating the 6 base pairs of the RBS, the 5 of the spacer, and the first base of the start codon. We begin with the 'native' RBS (BBa_K1383000), which is a known functional RBS sequence in methanogens and is typically used for creating synthetic parts. The RBS variants are created by making mutations on only one base at a time for every nucleotide different from the native (figure 2). We designed two additional RBS sequences based off the 16S rRNA data for M. maripaludis, termed theoretical 'perfect' and 'negative' (figure 2, #37 & #38, BBa_K1383001 (shown on this page) & BBa_K1383002, respectively). These RBS sequences were designed to have the theoretical greatest and worst possible affinities for the 16S ribosome. Figure 3 illustrates the specific sequence for the part described on this page, the 'native' RBS.
In an effort to expand synthetic biology research for Archaea, we have developed protein expression tools to facilitate fluorescence mediated detection of proteins. For our 2014 project we present 3 new BioBrick parts that iGEMers can use readily. All three of our parts are BioBrick compatible. Specifically, we constructed tools consisting of native/ synthetic Methanococcus RBS site(s) upstream of a gene encoding red fluorescent protein-mCherry. Prior to cloning the mCherry gene was codon optimized for expression in Methanoccocus (also, ensuring BioBrick compatibility in the design considerations). The BioBrick part- BBa_K1383001 consists of the theoretical strongest Methanococcus RBS site upstream of the mCherry gene. This fragment was inserted into pSB1C3 plasmid backbone using EcoRI and PstI restriction enzymes.
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000COMPATIBLE WITH RFC[1000]
| n/a | BBa_K1383001 (mCherry- Theoretical strongest RBS) |