Composite
Part:BBa_K1520024
Designed by: yue wu Group: iGEM14_Nanjing-China (2014-09-22)
Pcons2-rbs-lacI-Ter-Plac-rbs-luxI-Ter-Pcons2-rbs-luxR-Ter-Prlux-rbs-rfp-rbs-luxI-rbs-luxR-Ter
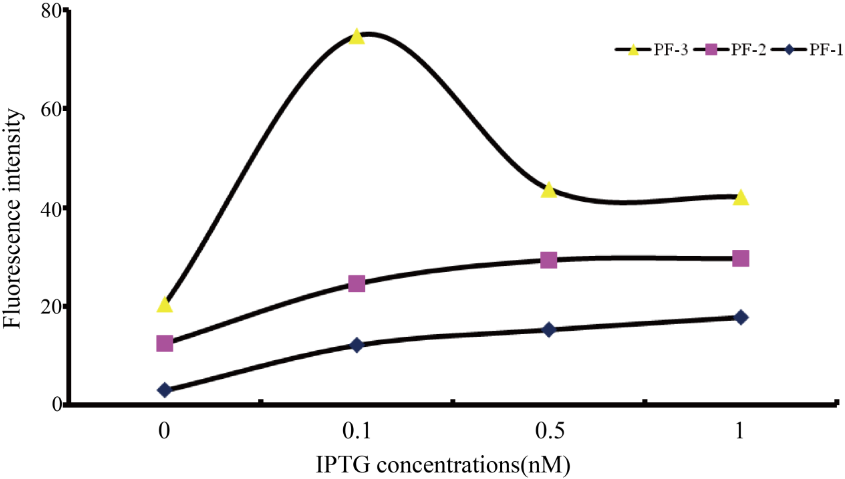
Plasmid in positive feedback verification: Pcons2-rbs-lacI-Ter-Plac-rbs-luxI-Ter-Pcons2-rbs-luxR-Ter-Prlux-rbs-rfp-rbs-luxI-rbs-luxR-Ter It is a positive feedback plasmid using luxI and luxR as key to feedback. Introducing IPTG will produce rfp, luxI and luxR, and luxI together with luxR will work back to enhance production of rfp.
The blue line in the picture shows the fluorescence intensity of the bacteria with this plasmid, which serves as a control in our experiment. The yellow line and purple line show the fluorescence intensity of the bacteria with different positive feedback plasmids.
Sequence and Features
Assembly Compatibility:
- 10COMPATIBLE WITH RFC[10]
- 12INCOMPATIBLE WITH RFC[12]Illegal NheI site found at 7
Illegal NheI site found at 30
Illegal NheI site found at 2380
Illegal NheI site found at 2403 - 21INCOMPATIBLE WITH RFC[21]Illegal BglII site found at 1208
Illegal BglII site found at 2222
Illegal BglII site found at 4818 - 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000INCOMPATIBLE WITH RFC[1000]Illegal BsaI.rc site found at 3358
[edit]
Categories
Parameters
//cds
//cds/transcriptionalregulator/activator
//cds/transcriptionalregulator/repressor
//function/cellsignalling
//function/regulation/transcriptional
//cds/transcriptionalregulator/activator
//cds/transcriptionalregulator/repressor
//function/cellsignalling
//function/regulation/transcriptional
| biology | |
| control | luxR, HSL |
| direction | Forward |
| ec_num | none |
| function | Mixed |
| kegg | none |
| ligands | HSL |
| n/a | constitutive promoter family member |
| negative_regulators | |
| o_h | |
| o_l | |
| positive_regulators | 1 |
| protein | LuxR |
| swisspro | P12746 |
| tag | None |
| uniprot | P48445 |