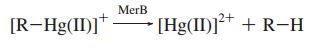Part:BBa_K1420002
merB, Organomercurial Lyase from Serratia marcescens
Overview
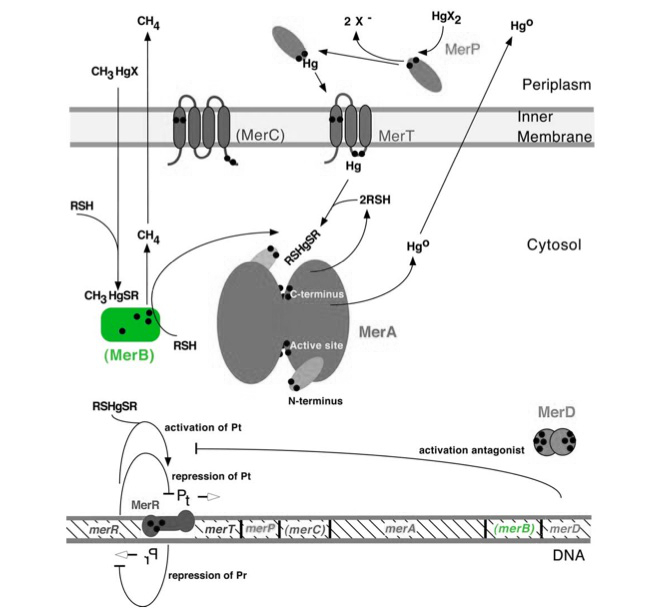
The gene merB (0.6Kb) encodes the carbon-mercury bond lyase MerB. The MerB enzyme along with its corresponding gene are highlighted in green in Figure 1. The lower portion of the Figure 1 shows the arrangement of mer genes in the operon, with merB located downstream of the transport and reductase genes, merP, merT and merA, respectively.
Figure 1. Model of carbon-mercury lyase operon. The symbol • indicates a cysteine residue. RSH indicates cytosolic thiol redox buffers such as glutathione. Figure 1 shows the interactions of MerB, in green, with mercury compounds and other gene products of mer operon. (Figure adapted from "Bacterial mercury resistance from atoms to ecosystems." 1)
Molecular Function
The merB gene is often found immediately downstream of merA, and is essential for the detoxification and bioremediation of organic toxic mercury compounds in congruence with merA.1 The merB protein is a lyase that catalyzes the breaking of carbon-mercury bonds through protonolysis of toxic mercury compounds, such as methylmercury (Scheme 1).2 This produces the less toxic and less mobile Hg2+ which is then completely volatilized to Hg0 when acted upon the enzyme merA.1
Scheme 1. MerB Catalyzed Reaction (R= alkyl, aryl)
Structure and Mechanism
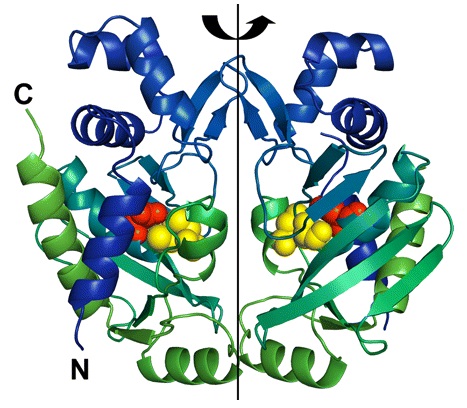
Enzyme MerB forms a dimer by a 2-fold pseudosymmetry around the depicted rotation axis in Figure 2.3 The two subunits are color coded from blue to green distinguishing the amino-terminal end of the protein from the carboxy-terminal, respectively. Active site residues are color coded as well using van der Waals sphere representation: residues Cys-96 and Cys-159 are in yellow, and residue Asp-99 is in red. The structure of mercury-bound MerB and free MerB is extremely similar with the exception of the mercury ion.3 The mercury is bound to MerB by two sulfurs from the Cys-96 and Cys-159. An oxygen from a water molecule is also involved, binding to the mercury atom.3 On the other hand, the active site residues for the free MerB are completely inaccessible.3
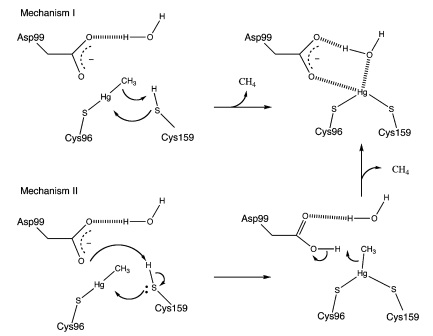
MerB can catalyze the protonolysis of carbon mercury bonds for a number of organomercurials, accelerating the rate of this reaction up to 107 times the rate of spontaneous abiotic decay.4 Two proposed mechanisms for how this is accomplished is displayed in Figure 3.4 Both mechanisms assume that the methylmercury substrate forms an initial bond with Cys96 and that the Cys159 site is protonated. In Mechanism 1, Cys159 protonates the methyl carbon and forms a covalent bond with Hg(II). In Mechanism 2 however, Cys159 donates a proton to Asp99 before forming a bond with Hg(II). Asp99 is then responsible for protonating the methyl group.
Figure 2. Structure of MerB enzyme. MerB is a dimer with 2-fold pseudosymmetry. The amino-terminal and carboxy-terminal are color coded blue and green, respectively. In addition, the active site residues Cys-96 and Cys-159 are in yellow, and residue Asp-99 is in red. (Figure adapted from "Enzyme Catalysis and Regulation: Crystal Structures of the Organomercurial Lyase MerB in Its Free and Mercury-bound Forms: Insights into the Mechanism of Methylmercury Degredation." 3)
Figure 3. Proposed mechanisms of MerB catalysis for the Hg-C protonolysis reaction. Both of these proposed mechanisms display how methyl group of methylmercury is protonated, producing Hg(II). Mechanism 2 differs from Mechanism 1 by utilizing Asp99 for protonation of the methyl group with the hydrogen attached to the sulfur from Cys159. (Figure adapted from "Mechanism of Hg-C Protonolysis in the Organomercurial Lyase MerB." 4)
merB Experimental Results
The function of MerB is to convert methylmercury to Hg(II). This functionality was tested in both silica gel encapsulated Escherichia coli K12 cells and unencapsulated E. coli cells. Both encapsulated and unencapsulated E. coli K12 containing the “pBBRBB::mer were compared to three negative controls; encapsulated and unencapsulated “E. coli K12 containing the pBBRBB::gfp plasmid and LB media(abiotic). Negative controls were used to determine the amount of methylmercury absorbed by beads and the natural degradation rate of methylmercury in un-inoculated media. The experiment was conducted by adding methylmercury chloride (0.1M) to 7 mL of LB for a starting concentration of 1 mg/L methylmercury chloride. Silica gel beads material (0.5mL) was used to encapsulate both “pBBRBB::mer” and “pBBRBB::gfp”, both the unencapsulated and encapsulated samples were inoculated at an OD ~ 0.08. Time points were then taken to determine a rate of degradation of methylmercury. Samples were taken at time points of 1, 2, 4, 8, and 24 hours, and were diluted a million-fold before taking measurements. The samples were analyzed using a Tekran model 2700 Automated Methyl Mercury Analyzer using EPA method 1630 without distillation. This is a highly sensitive and ultra-stable cold vapor atomic fluorescence spectrometry (CVAFS) Hg detector. All quality assurance and quality control measures were taken as outlined in EPA method 1630. All MeHg standards (ongoing precision recoveries) were within the acceptable range averaging 96%.</p>
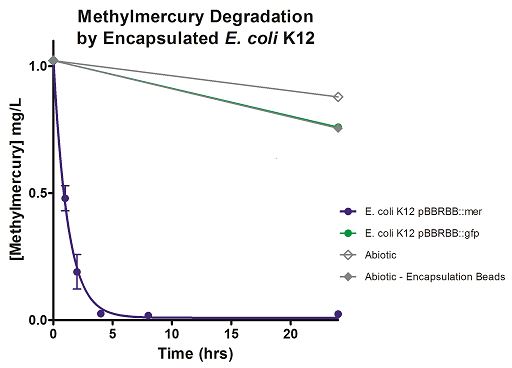
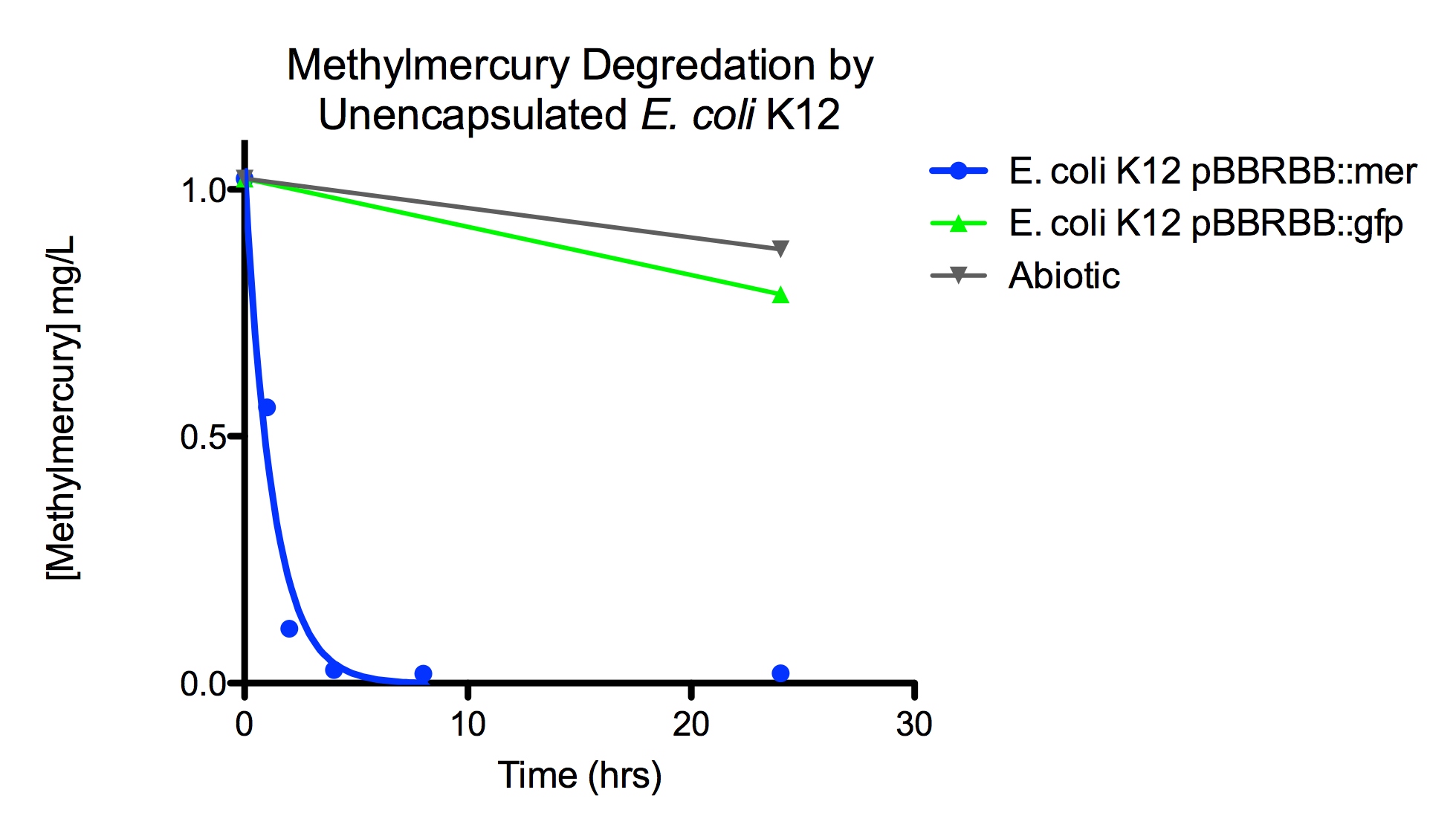
The final measured results of the methylmercury rate test are depicted in Figure 4 illustrating the degradation of methylmercury in encapsulated bacteria and “Figure 5” showing the degradation of methylmercury using unencapsulated bacteria. The data show that negligible amounts of methylmercury are degraded in abiotic solutions. The data suggest that the negative control: K12 strain containing the pBBRBB::gfp, had a more significant decrease in concentration of methylmercury over time when compared to the abiotic control, but is still negligible in comparison to the E. coli K12 containing the pBBRBB::mer. The mer operon, is seen to remove methylmercury from the system almost completely after only 8 hours.
From the data, it can be extrapolated that the presence of merB in our pBBRBB::mer construct facilitates the bioremediation and degradation of mercury. These results confirm the proposed mechanism of MerB and its function in the cleavage of carbon groups from methylmercury reducing it to Hg(II).
Figure 4. Monitoring the levels of mer operon methylmercury bioremediation. The graph displays that abiotic, abiotic with encapsulation beads, and the E. coli K12 strain containing the pBBRBB::gfp control bioremediate methylmercury from the environment at a much slower rate than the E. coli K12 containing the pBBRBB::mer, which can remove methylmercury from the system almost completely in approximately 8 hours.
Figure 5. Monitoring the levels of mer operon methylmercury degradation. The graph displays that abiotic and E. coli K12 strain containing the pBBRBB::gfp control degrad methylmercury from solution at much slower rates than the E. coli K12 containing the pBBRBB::mer, which can degrad methylmercury from the system almost completely in 8 hours.
References
1. T. Barkay et al (2003). "Bacterial mercury resistance from atoms to ecosystems." FEMS Microbiology Reviews 27: 355-384.
2. V. B. Mathema et al (2011). "Bacterial mer operon-mediated detoxification of mercurial compounds: a short review". Archives of Mictobiology 193: 837-844.
3. J. Lafrance-Vanasse et al (2009). "Enzyme Catalysis and Regulation: Crystal Structures of the Organomercurial Lyase MerB in Its Free and Mercury-bound Forms: Insights into the Mechanism of Methylmercury Degredation."The Journal of Biological Chemistry 284: 938-944.
4. J. M. Parks et al (2009). "Mechanism of Hg-C Protonolysis in the Organomercurial Lyase MerB."Journal of American Chemical Society 131: 13278-13285.
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25INCOMPATIBLE WITH RFC[25]Illegal NgoMIV site found at 415
- 1000COMPATIBLE WITH RFC[1000]
//cds
| None |