Part:BBa_J179000
This part is an empty backbone vector for the usage in Bacillus subtilis. It integrates in the amyE locus and can be selected with Chloramphenicol (cat gene). It has a ampicillin resistance for cloning in E.coli. This backbone is a BioBricked version of the B. subtilis vector pDG1662 ([http://www.ncbi.nlm.nih.gov/pubmed/8973347 Guérout-Fleury et al.])
This version does not contain AgeI nor NgoMIV restriction sites, therefore is RFC25-compatible.
This BioBrick is part of the [http://2012.igem.org/Team:LMU-Munich/Bacillus_BioBricks BacillusBioBrickBox] developed by the LMU-Munich 2012 iGem Team.
For handling of B. subtilis vectors, please see here.
The transformation into B. subtilis is explained here.
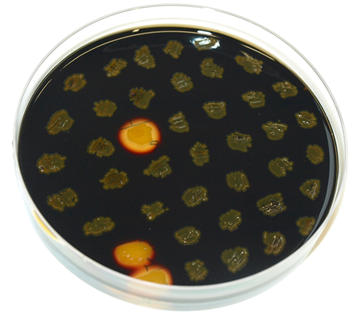
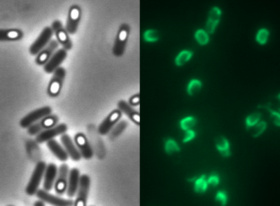
To check for integration of this vector into the amyE locus, the starch test can be used. The colonies obtained from the transformation into B.subtilis were streaked on an LB-Agar plate with 5 mg/L chloramphenicol and onto a starch plate (also W168 as a control) and incubated at 37°C over night. The starch plate is then covered with Lugol's iodine. Colonies without a bright surrounding are correct. We tested this vector with various inserts, for example the final construct of our Sporobeads with GFP. The starch test shows that the insertion in in the amyE locus and fluorescence microscopy reveals the functionality of the construct.
Also the iGEM-Team from [http://2012.igem.org/Team:Groningen/international_cooperation Groningen 2012] verified that this backbone works just fine.
//chassis/prokaryote/bsubtilis
//chassis/prokaryote/ecoli
//plasmid/chromosomalintegration
//plasmidbackbone/assembly
//plasmidbackbone/operation
| chassis | B. subtilis + E. coli |
| copies | |
| origin | |
| resistance | A(E. coli) C(B. subtilis) |