Part:BBa_K1152007:Experience
This experience page is provided so that any user may enter their experience using this part.
Please enter
how you used this part and how it worked out.
General application of BBa_K1152007
pJS01 was amplified out of four fragments via Gibson Cloning: ccdB, the C-domain of TycC2 and the indC-gene, as well as a backbone. We verified the genotype by colony-PCR, restriction digest with various restriction enzymes, among them EcoRI and PstI, and sequencing of the assembly overhang regions. With the RFC100 standard, we propose a framework for NRPS-based, customized peptide synthesis. This includes the possibility to tag the desired Non-Ribosomal Peptides with Indigoidine. Using this helper-construct eased our work in bringing the proof of principle for the functionality of the Indigoidine-tag, as by replacing the ccdB with the NRPS modules of interest, re-ligation background is significantly decreased. This construct stands as representative of the various fusion-peptides with Indigoidine-tag that we synthesized during our project.
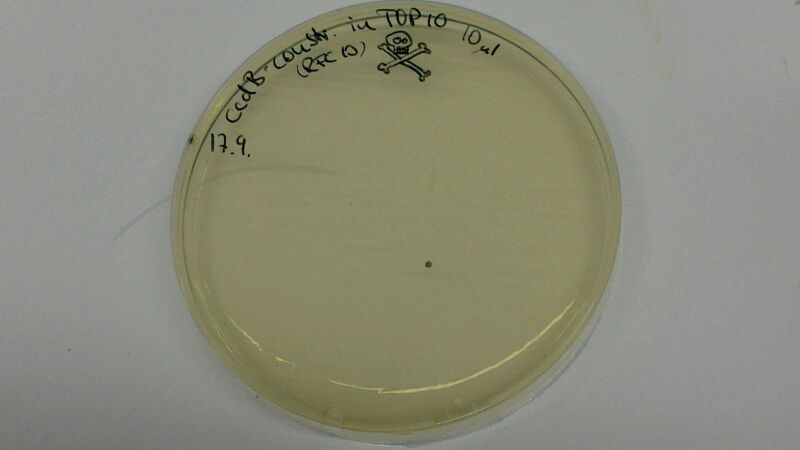
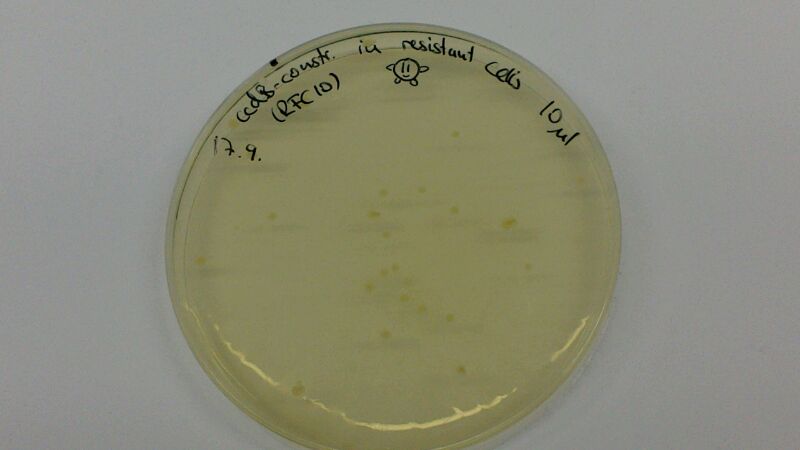
(Figure 1 & 2: In the left picture, one can see the result of a transformation of ccdB-resistant cells with pJS01. On the plate where non-resistant TOP10-cells were transformed with ccdB-Ind (figure 2), hardly any cells are growing. Since cells were not induced with IPTG, culutres did not turn blue)
The comparison of the figures 1 and 2 attests the significant decrease of background when using the ccdB-helper construct.
Concrete examples of Indigoidine-tagged peptides
Valine-Indigoidine
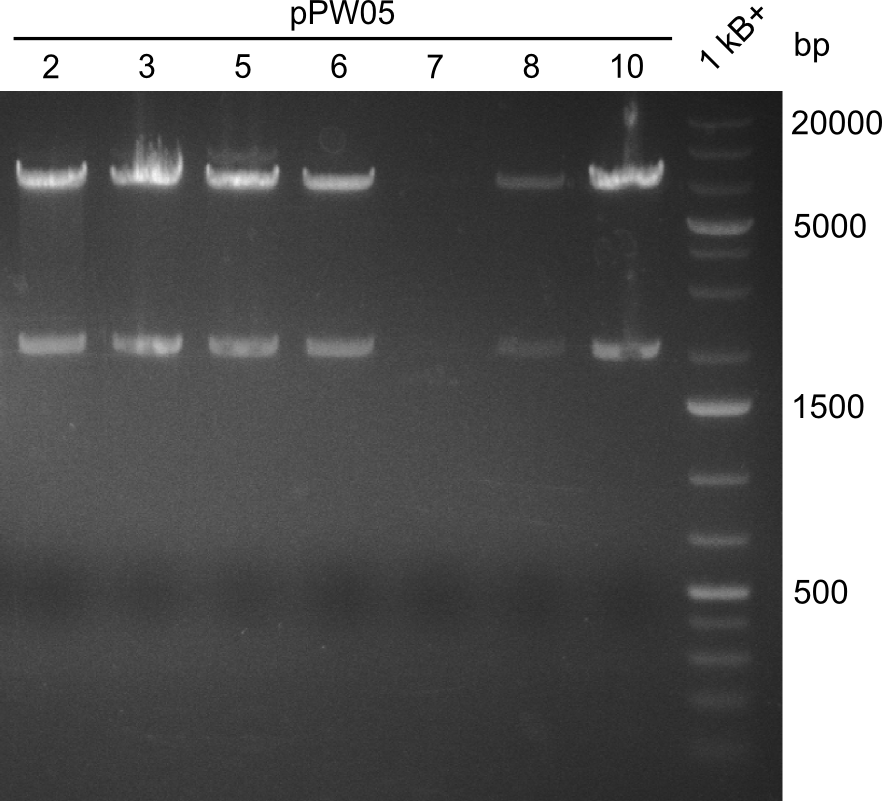
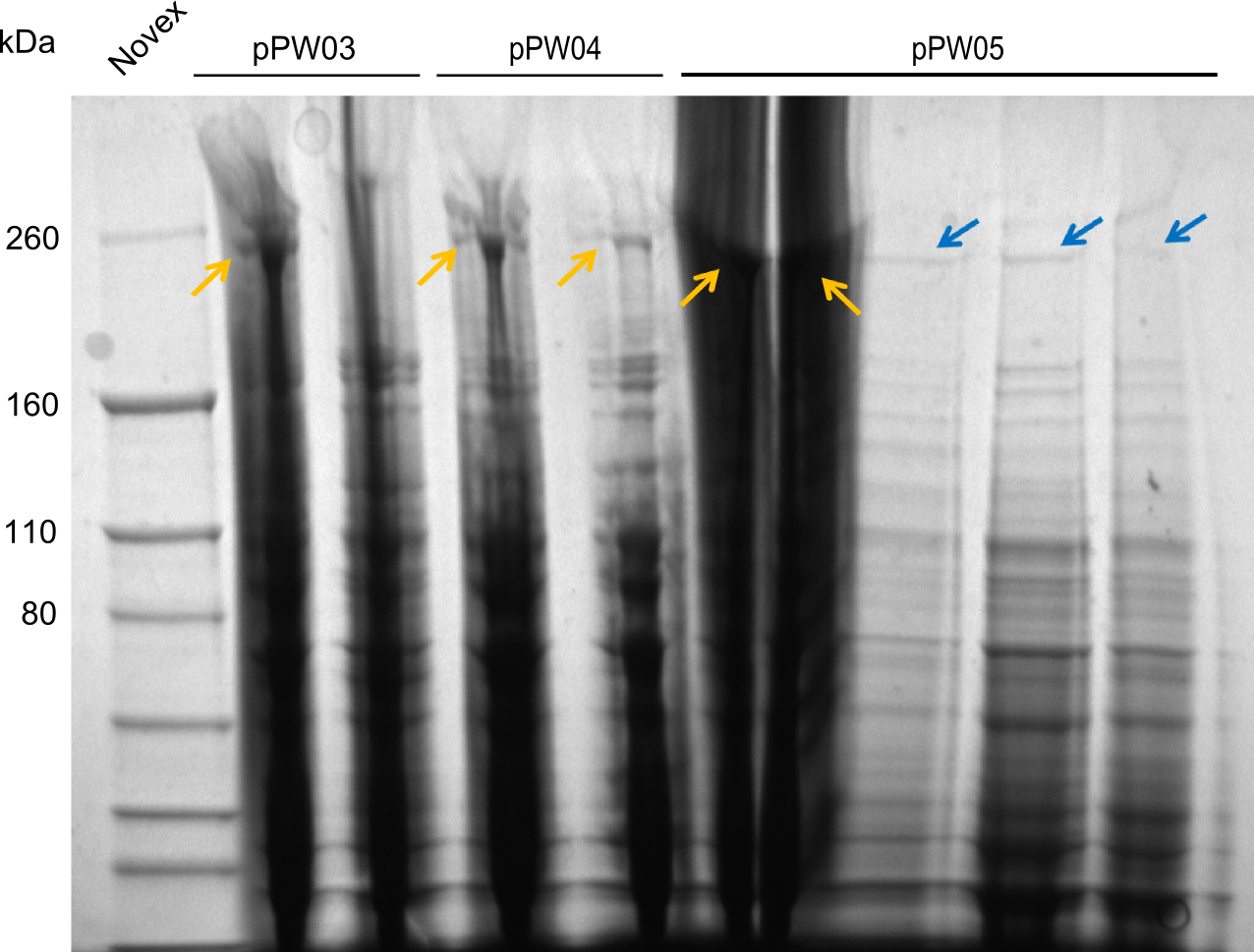
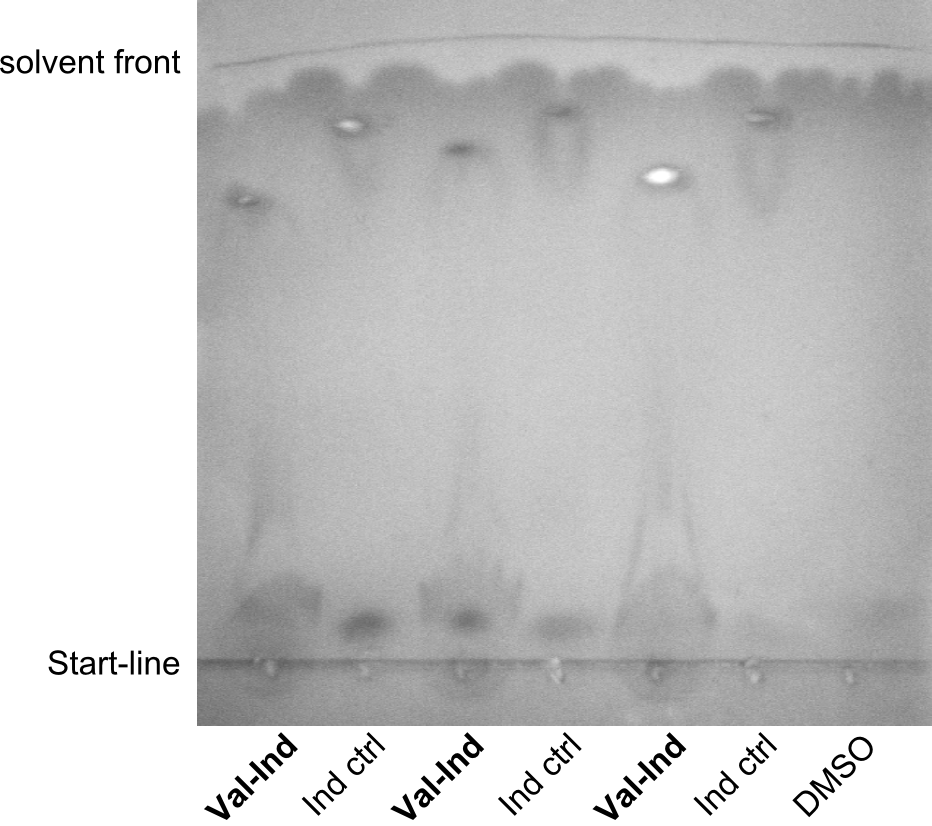
After amplification of pJS01 and TycC4dC (which is a module from B. parabrevis), we assembled the Val-Ind-Synthetase using Gibson Cloning. In this approach, ccdB was replaced by the Tyrocidine-module. We verified the genotype by colony-PCR, restriction digest with various restriction enzymes, among them EcoRI and PstI, and sequencing of the assembly overhang regions. Furthermore, SDS-PAGE was performed after induction of cells in order to prove expression – the expected band at 242 kDa was clearly visible after Coomassie staining. Finally, after purification of the NRP, TLC on silica-gel with Dichloromethane as mobile phase, was performed to prove the functionality of the expressed NRPS.
(Figures 3, 4 & 5: Verification of the genotype, the expression and the functionality of BBa_K1152006)
In restriction digest, as one can see in figure 3, the cutting pattern was as expected. For restriction digest with EcoRI and PstI, as expected two bands show up, one at around 2000 bp (backbone) and one at around 7000 bp (insert).
In the SDS-PAGE, expression of the NRPS could be verified, as the expected band at 242kDa is clearly visible after induction with IPTG (see figure 4). To prove that the Val-Ind NRPS is in fact functional, we purified the synthesized NRP and analyzed its properties by comparative TLC with an Indigoidine control. As one can see in figure 5, the migration behavior of the synthetic fusion peptide is significantly different from the one of Indigoidine. The samples for Val-Ind tested on TLC were extracted from different clones, hence reproducibility by biological replicates could be proven.
Ornithine-Valine-Indigoidine
Having shown the functionality of pJS01 in the assembly of pPW05, we assembled further constructs with an Indigoidine-tag. This plasmid (pPW06) was examined for functionality by TLC only, as this is the very charm of the standard we propose: The success of an assembly is visible to the unaided eye.
(Figure 6: Comparative TLC of Orn-Val-Ind with Indigoidine-control shows altered migration behavior for the tagged peptide)
By comparative TLC on silica-gel with Dichloromethane as liquid phase, we could show that the Orn-Val-Ind-Synthetase which is encoded by pPW06 is in fact functional. Furthermore, we could demonstrate that the ccdB-helper-plasmid approach is effective and straightforward.
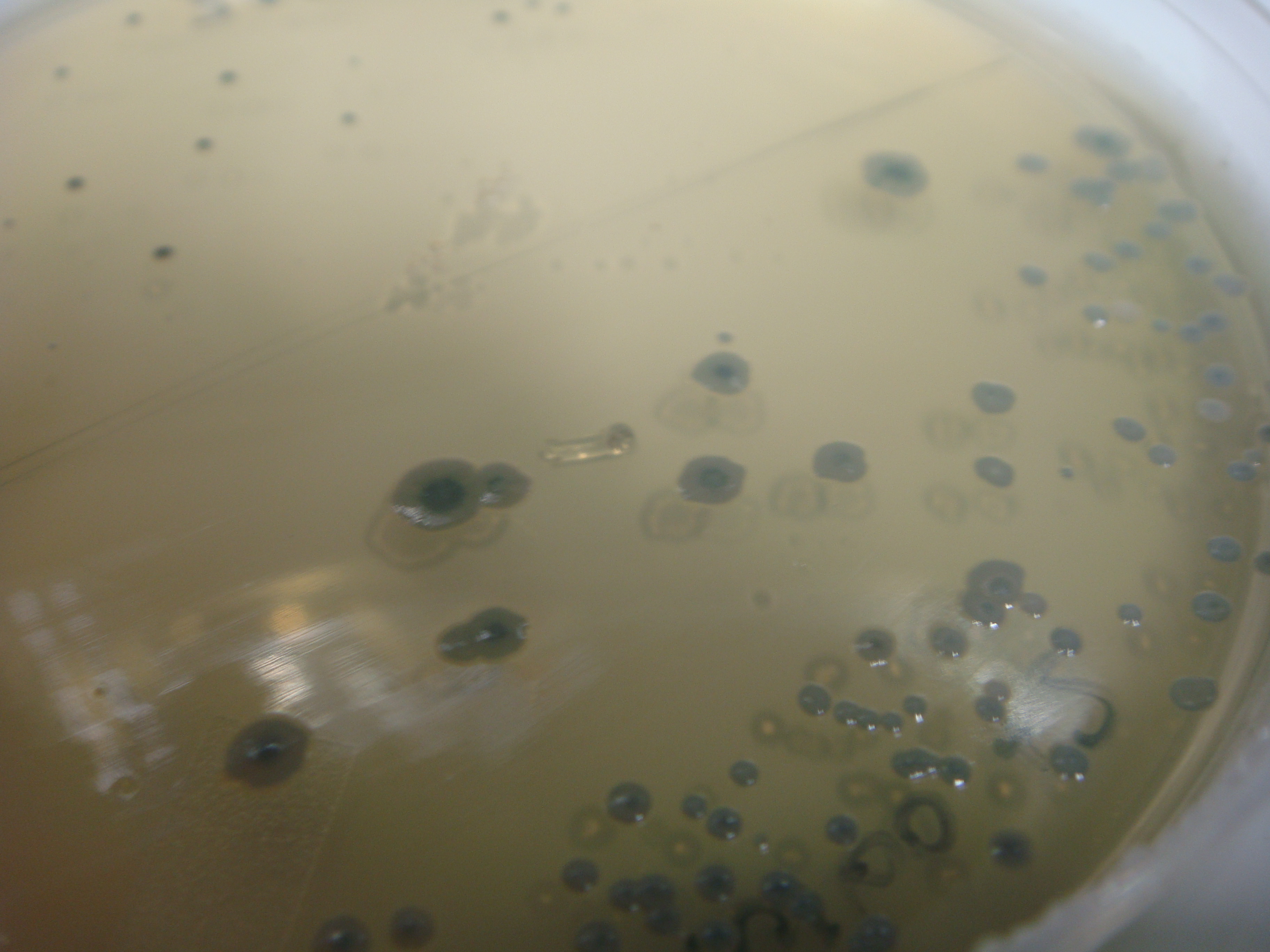 (Figure 7: Showcase of blue colonies after IPTG induction)
(Figure 7: Showcase of blue colonies after IPTG induction)
User Reviews
UNIQ8bf03eee87a63ca0-partinfo-00000000-QINU UNIQ8bf03eee87a63ca0-partinfo-00000001-QINU