Part:BBa_K1033000
Tyrosine ammonia-lyase (TAL) with RBS
Tyrosine ammonia-lyase (TAL) is an enzyme which catalyzes the formation of p-coumaric acid (aka p-hyroxycinnamic acid) from tyrosine. It belongs to the family of ammonia-lyases, enzymes that catalyze the deamination of amino acids [1]. P-coumaric acid is an important precursor in many metabolic pathways. TAL also has a secondary function as a phenylalanine ammonia-lyase (PAL) which catalyses the formation of cinnamic acid. This bacterial version have proved to have a hundred fold higher preference for tyrosine as a substrate, as opposite to many eukaryotic versions. [2].

Applications
This biobrick could have lots of applications. First it can by itself produce the antioxidant p-coumaric acid from tyrosine. The enzyme could also be mutated so that it catalyzes the formation of trans-cinnamic acid from phenylalanine.
P-coumaric acid is also needed as sa precursor for many metabolites in the phenylpropanoid pathway. Examples of these are resveratrol that we have worked on. (link) Also many other compunds like flavanones, lignins and anthocyanins can be produced from p-coumaric acid. With this enzyme, we have opened up the phenylpropanoid pathway for bacteria.
How to characterize it
P-coumaric acid shows no visible color, so one has to characterize it with the help of methods like spectrophotometry and chromatography.
Characterization data from Uppsala iGEM 2013
Summary
Tyrosine ammonia lyase (TAL) from rhodobacter sphaeroides was obtained from J.Conrado et al[4] We biobricked TAL with the ribosome binding site B0034 and overhangs in a single pcr. We also made a version with 6-HIS-tag for enzyme expression analysis. TAL was also mutagenized to remove illegal Not1 site. We verify all of our genetical constructs with sequencing.
We have expressed TAL in e-coli DH5alpha and E-coli nissle, a probiotic e-coli obtained from Trieste iGEM 2012. TAL will also be characterized in lactobacillus, by transforming the construct with our shuttle vector. This construct can also be used to produce the precursor for example our resveratrol.
We managed to clone out and biobrick tyrosine ammonia lyase and verify the biobrick by sequencing. Also we did succeful characterization on this part, showing that it works as expected. We managed to express our enzyme and detect it in a western blot, and also detect our metabolite in both spectrophotometry and hplc. The biobrick was characterized in e-coli d5halpha and e-coli nissle. Biobrick We succeded in the cloning and sequencing of our biobrick, Tyrosine ammonia lyase from rhodobacter sphaerides with the RBS B0034 that should work in various organisms, lactobacillus and e-coli. Sequencing was done at GATC biotech and Uppsala Genome center using sanger sequencing. Tyrosine ammonia lyase with rbs: BBa_K1033000.
Western blot We also succeeded in expressing the enzyme tyrosine ammonia-lyase (TAL) in e-coli. To enable the detection of this protein by anti-his antibodies, 6-histidine tags was incorporated in the sequence. This way we could detect our enzyme with anti-his antibodies. We expressed our protein with a multipromotor working in both lactobacillus and e-coli. This way, we can easily transfer TAL to lactobacillus later on. The size of our protein was calculated using ProtParam[5], 54.9 kDA.

Figure 1:SDS-page and western blot. Expression of Tyrosine ammonia lyase with constitutive promotors. The negative control is empty, showing that there is no natural protein in e-coli with the same attributes.
1.Positive control
2.TAL with Cp8 promotor
3.TAL with J23110 promotor
4.Negative control
Spectrophotometry
As the next step, we have made a spectrophotometric assay of our metabolite p-coumaric acid produced by e-coli. By using n-octanol and a two-phase extraction, we were able to extract our metabolite from the lb medium and bacteria. This way we could characterize our biobrick on spectrophotometer. (protocol link)



Figure 2:Absorbance spectra of extracts collected from bacterial cultures. Samples were collected 21 h and 48 h after 30 °C incubation. Negative control is an extract from a strain with no TAL gene on a transformed plasmid. The positive control is an extract on a culture of the same strain as the negative control but with added p-coumaric acid to a concentration of 500 ”M before extraction. High pressure liquid chromatography
To further prove p-coumaric acid production in e-coli, we did a hplc of our bacterial culture. We managaed to detect our metabolite at around 9 minutes. (Protocol link)
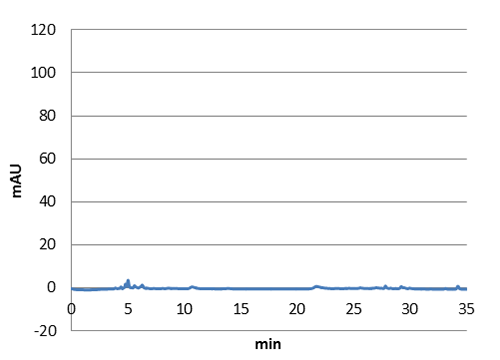
Figure 3: E-coli culture injected to the hplc without our biobrick tyrosine ammonia lyase. Here we can see that there is originally no peak at 10 around minutes.

Figure 4. Graph showing the HPLC result of a sample standard with p-coumaric acid

Figure 5. Graph showing the HPLC result of a sample prepared from e coli expressing tyrosine ammona lyase. In this graph, the promotor J23101 is used. Reverse phase HPLC with a C18 matrix was used. The peak for p-coumaric acid can be seen ~10min, as shown by the standard sample below.
References
1.Zhixiong Xue, Michael McCluskey, Keith Cantera, F. Sima Sariaslani, Lixuan Huang (2007) Identification, characterization and functional expression of a tyrosine ammonia-lyase and its mutants from the photosynthetic bacterium Rhodobacter sphaeroides. J Ind Microbiol Biotechnol 34:599-604
2. J.A. Kyndt, T.E. Meyer, M.A Cusanovich, J.J. Van Beeumen (2002) Characterization of a bacterial tyrosine ammonia lyase, a biosynthetic enzyme for the photoactive yellow protein. FEBS Letters 512 240-244
3. Robert J. Conrado et al, DNA guided assembly of biosynthetic pathways promotes improved catalytic effiency. Nucleic Acids Research , 2012, Vol 40 NO 4, 1879-1889
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25INCOMPATIBLE WITH RFC[25]Illegal NgoMIV site found at 45
Illegal NgoMIV site found at 877
Illegal AgeI site found at 140
Illegal AgeI site found at 306 - 1000INCOMPATIBLE WITH RFC[1000]Illegal BsaI site found at 1335
//chassis/prokaryote/ecoli/nissle
//chassis/prokaryote/lactobacillus
//collections/probiotics
//collections/probiotics/production
| None |
