Part:BBa_K1094401
MamC-eGFP fusion
MamC (BBa_K1094001) from Magnetospirillum magnetotacticum (MS-1) tagged with enhanced green fluorescent protein (eGFP, BBa_K1094400). Between the parts a glycine linker (10 glycine residues) is added. The gene product can be used to detect localization of MamC in MS-1.
The two parts was cloned together using classical restriction digestion cloning in pBBR1MCS-2. The composite part was later PCR amplified and cloned into the expression vector pJAM1786 via pDONR207 in E. coli, by using the Gateway system by Invitrogen. Colony PCR and gel electrophoresis was performed to ensure the insert was in the vector.
The restreaks of 6 colonies were inoculated into liquid culture and grown overnight alongside three E. coli cultures containing a non-expression plasmid with a non-fluorescent insert (MamC-pSB1C3). Fluorescence was measured in an opaque ELISA reader plate. The following dilutions were made: 1X, 2X, 5X, 10X and 50X. OD(600nm) was measured to ensure similar cell density. Excitation was carried out at 485 nm and emission was collected at 535 nm.
The mean values of the fluorescence can be found in the table below.

(The raw data can be found here)
All dilution showed significantly more fluorescence than the control cultures except for the 50X dilution. The mean of the difference to the control cultures is shown as a function of dilution below.
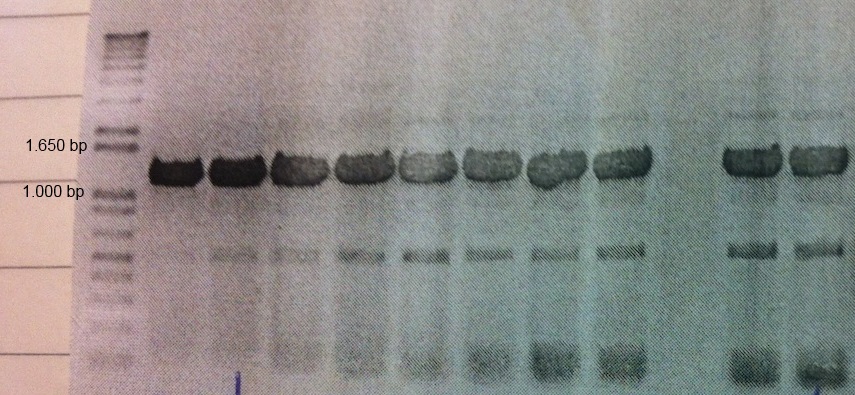
A Western blot of the eGFP-pJAM1786 cultures was made with anti-GFP as primary antibodies. The eGFP-pJAM1786 samples give rise to band of the same size as a GFP positive control (see figure below). Unfortunately, the size standard was lost during the transfer to the membrane.

Fluorescence- and confocal microscopy was carried out.
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25INCOMPATIBLE WITH RFC[25]Illegal NgoMIV site found at 169
Illegal NgoMIV site found at 253 - 1000COMPATIBLE WITH RFC[1000]
| None |