Part:BBa_K1100011
B0015 terminator efficiency estimation device (with Csy4 system)
In this device, we test the terminator efficiency in the consideration that the terminator is influenced by the downstream sequence. The csy4 cleavage site is inserted just after the terminator(BBa_B0015).
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25INCOMPATIBLE WITH RFC[25]Illegal NgoMIV site found at 448
- 1000INCOMPATIBLE WITH RFC[1000]Illegal SapI site found at 164
Background
Csy4 is a member of CRISPR pathway discovered in Pseudomonas aeruginosa. It is a single endoRNase that recognizes and cleaves a 28-nucleotide repetitive sequence and produces stable transcripts with a 5’hydroxylgroup, which can eliminate unwanted interactions between 5’UTRs and translational elements such as RBSs units to standardize the expression of the elements. ( Lei Qi et al, 2012).The cleavage site can be curtailed to 16nt proved by in vitro data (Rachel E. Huarwitz et al, 2010)
Figure. 1. The CRISPR RNA-processing system facilitate engineering of standard genetic elements in various contexts( Lei Qi et al, 2012).
Design
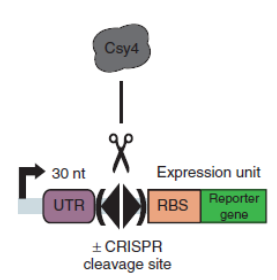
Csy4 system is introduced into the terminator efficiency test assay and used optimized sfGFP to test the PoPs of Ptet (BBa_R0040), to approximately estimate the input of BBa_B0015. Then we tested the output of BBa_B0015, with Csy4 insulator to avoid the RNA interference.
Figure A. Construction for termination efficiency estimation. We used optimized sfGFP to test the PoPs of Ptet (BBa_R0040), to approximately estimate the input of BBa_B0015. Then we test the output of BBa_B0015, with Csy4 insulator to avoid the RNA interference to sfGFP translation.
Experimental Data (Quantitative Measurement)
We get the device measured by flow cytometer by testing the expression of sfGFP.
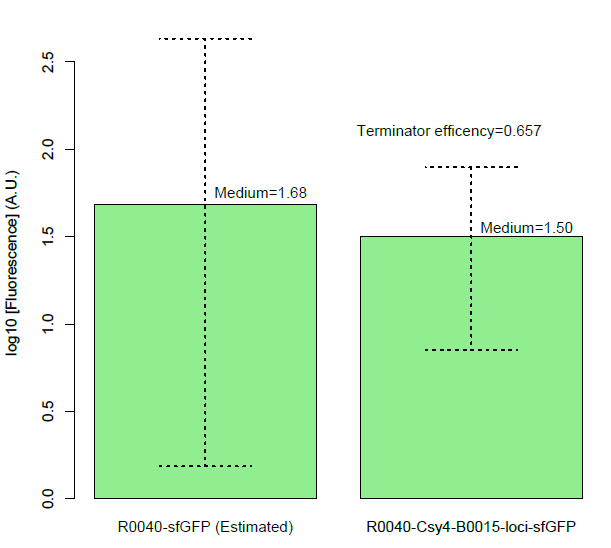
Figure B, Histogram of the expression of output of terminator, measured by sfGFP fluorescence by flow cytometer. We assembled the promoter BBa_R0040 and the Csy4 protein with the terminator BBa_B0015. And we used the sfGFP protected by a insulator to measure the leakage rate of the terminator.
Then, we found a decrease of terminator efficiency to only 0.657 (Figure C), due to the downstream interference, largely inconsistent with the previous study that showed that BBa_B0015 is a strong terminator, with 0.984[CC], 0.97[JK],forward efficiency, of which the rrnB_T1 (B0010) part also has 97.27% forward efficiency (G. Cambray, et al., 2013).
Figure C, Measurement of BBa_B0015 efficiency: column of the median values of fluorescent expressions with/without the terminator and the interaction between the csy4 and the 16-nt cleavage site. (Left: Input/Right: Output) The error bar represents two quartiles.(Up:75%, Down:25%) The terminator efficiency is (1-output value/input value).
Reference:
Haurwitz R E, Jinek M, Wiedenheft B, et al. Sequence-and structure-specific RNA processing by a CRISPR endonuclease[J]. Science, 2010, 329(5997): 1355-1358. Qi L, Haurwitz R E, Shao W, et al. RNA processing enables predictable programming of gene expression[J]. Nature biotechnology, 2012, 30(10): 1002-1006. Cambray G, Guimaraes J C, Mutalik V K, et al. Measurement and modeling of intrinsic transcription terminators[J]. Nucleic acids research, 2013, 41(9): 5139-5148.
| abs | B0034 |
| biology | |
| chassis | E.coli |
| control | aTc, tetracyline |
| direction | Forward |
| efficiency | 1 |
| emission | 510nm |
| excitation | 485nm |
| negative_regulators | 1 |
| o_h | |
| o_l | |
| positive_regulators | |
| protein | sfGFP |