Part:BBa_K1100141
J23101-ALeaderT
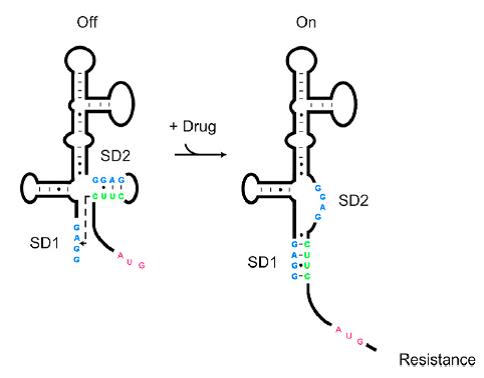
As shown in Figure 1, the 75nt RNA sequence of Aleader contains two SD sequences (ribosome binding sites) and an anti-SD sequence (CUUC) which can complementarily pair with either of the SD sequences. In the absence of aminoglycosides, anti-SD pairs with SD2. The binding of ribosomes to SD1 triggers the translation of a small peptide which stops at the stop codon ahead of SD2, and therefore inhibits the translation of the desired gene after SD2. When aminoglycosides (kanamycin for example) exists, it will induce a structural change of Aleader. The anti-SD sequence base-pairs with SD1, consequently unmasking SD2 for ribosomal binding, which results in the translation of the following gene.Many riboswitches have lower dynamic range and higher leakage rate compared with protein regulators. To reduce the basal level and enhance the dynamic range, while having no effect on the protein product, we designed a novel riboswitch called ALeaderT, following the principle of three-phase switch design.
We added a GGGS linker (dock III), a common neutral peptide linker, after the SD2 and ATG. Impressively, by RNA 2D-prediction, we found that dock III will capture antiSD2 sequence to form a stable stem loop, with characteristics of terminator, though neutral as an amino peptide. We suppose that the ALeaderT will have three different steady-state conformations, A “Translation off Termination off”, B “Translation on Termination off”, and C “Translation on Termination on”(Figure 2).
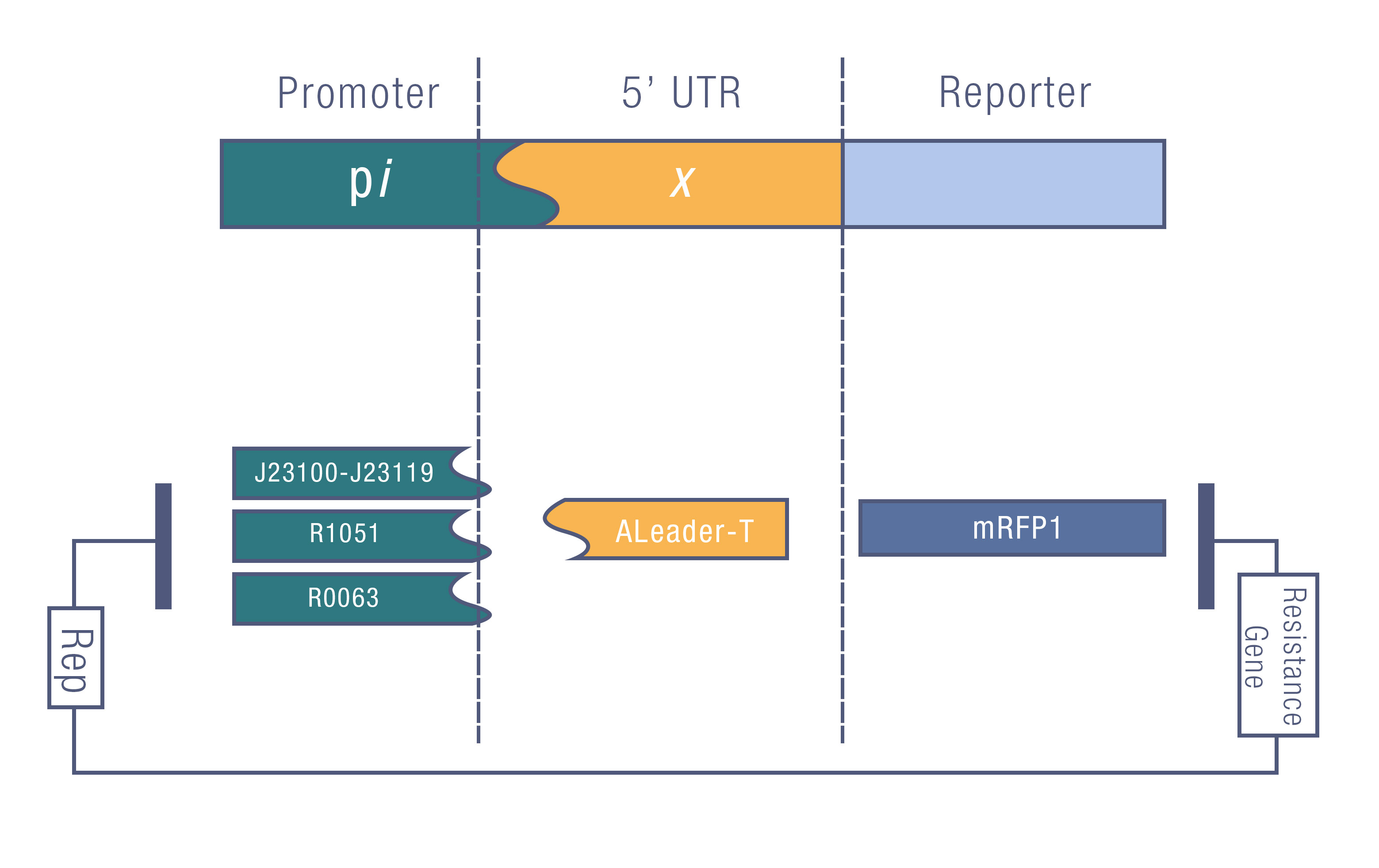
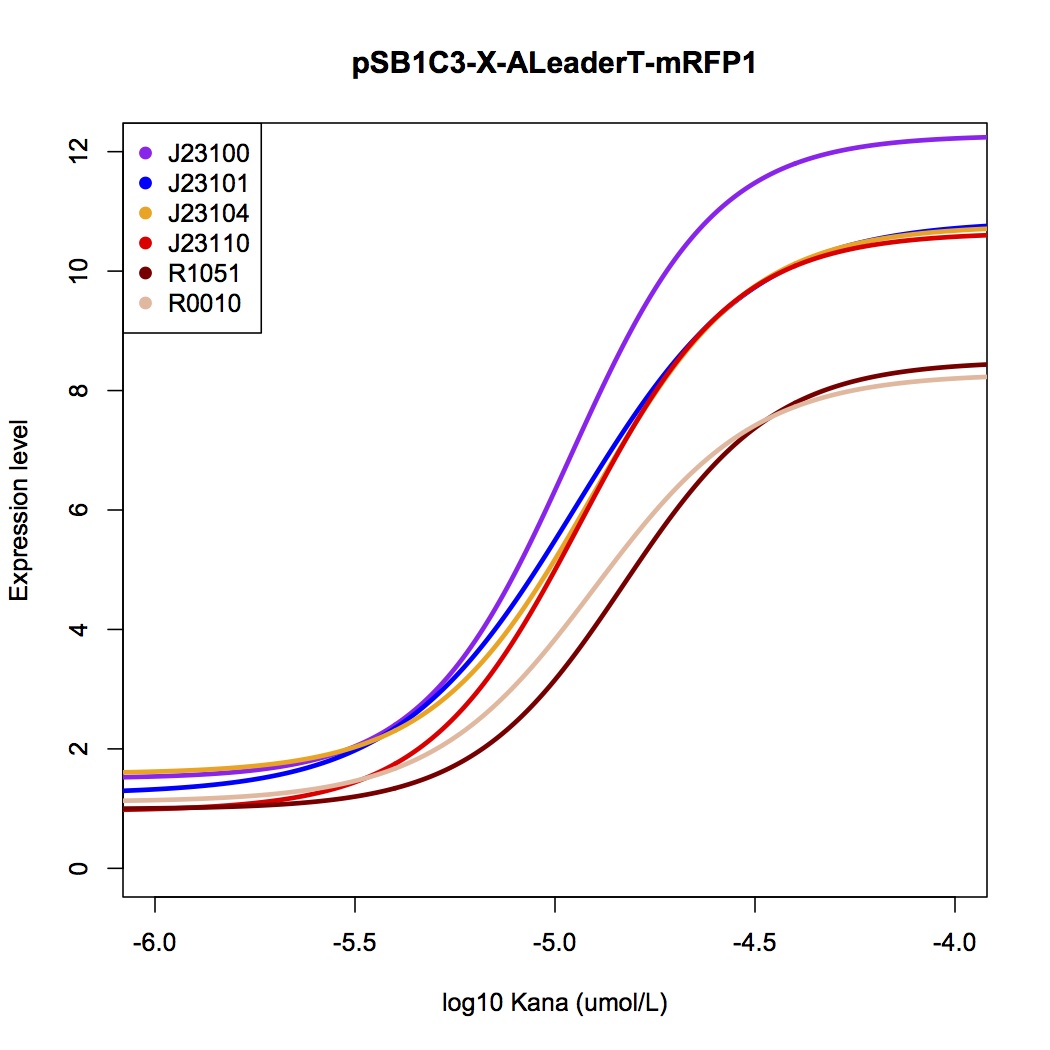
We design a series of experiments to test the function of ALeaderT with the reporter mRFP1 and a promoter library(Figure 3). It showed a very low minimal level and high dynamic range, consistent with the previous prediction. Moreover, it changed the hill function working curve into a square-like curve. Surprisingly, the ALeaderT also showed virtues of marvelous low noise in the system and minimal difference under different regulation. Therefore it will have its talent on the precisely regulation of RNA and protein expression.
We tested the ALeaderT’s function on translation control with the reporter mRFP1 and a promoter library measured by fluorescent.
It showed a very low basal level and high dynamic range, consistent with the modeling prediction(Figure 5,6). Marvelously, the ALeaderT is also much more standardized and robust than the wild type ALeader(Figure 5). In other words, with different promoters, AleaderT’s function varies insignificantly. Therefore it is talented on the precisely regulation of RNA and protein expression.

Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12INCOMPATIBLE WITH RFC[12]Illegal NheI site found at 7
Illegal NheI site found at 30 - 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000COMPATIBLE WITH RFC[1000]
| control | aminoglycoside antibiotics; Kanamycins |
| direction | Forwards |
| ligands | aminoglycoside antibiotics |
| positive_regulators | 1 |