Part:BBa_K322921:Experience
This experience page is provided so that any user may enter their experience using this part.
Please enter
how you used this part and how it worked out.
Applications of BBa_K322921
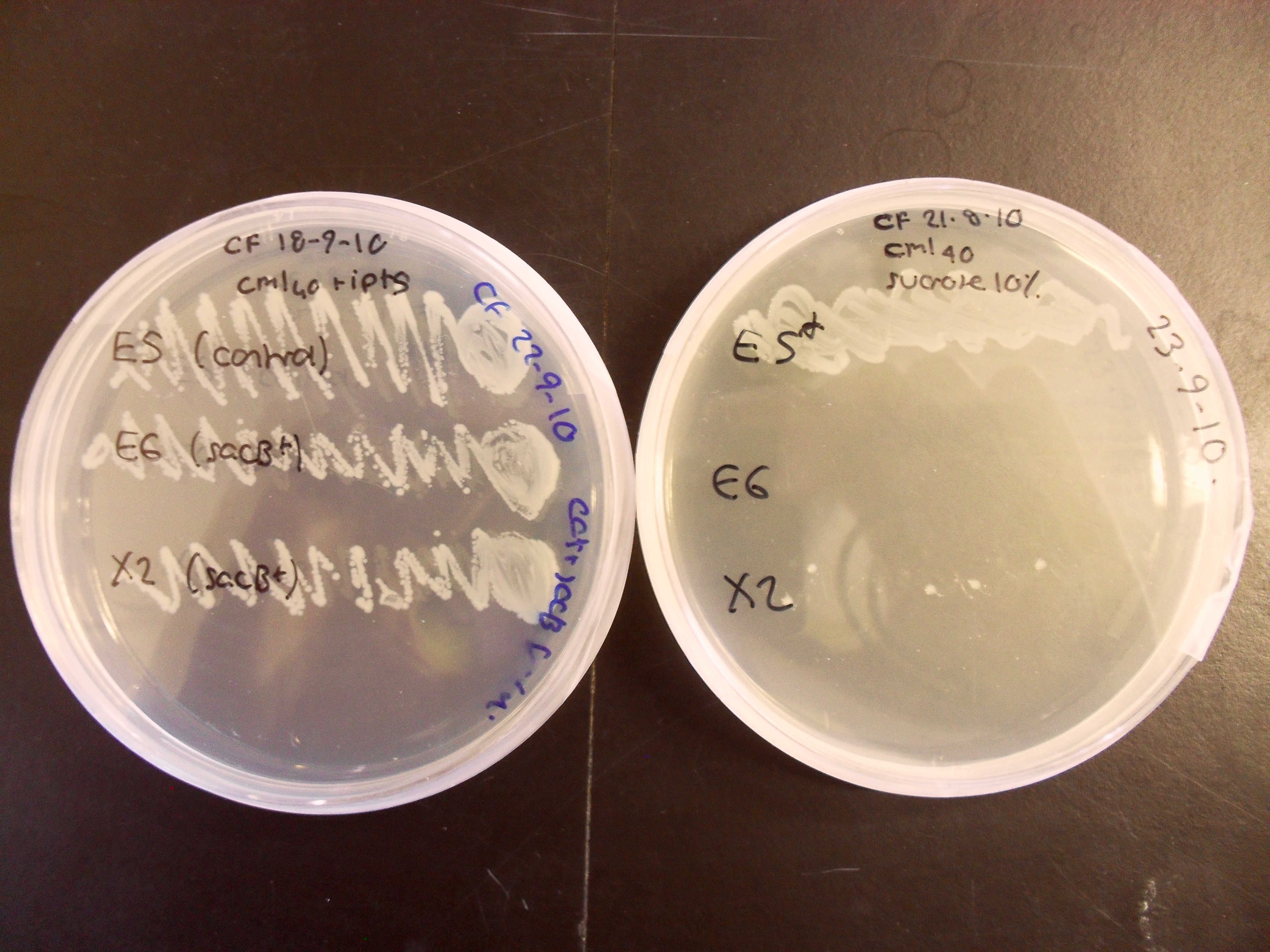
Characterisation of the activity of the sacB BioBrick:
The control strain which does not contain sacB has grown well on both plates, whereas the sacB-containing strain X2 grows poorly on 10% sucrose and E6 does not grow at all. This experiment confirms that sacB is effective as a negative selection marker within the BRIDGE protocol.
Further characterisation and preparation of improved constructs for sacB BioBrick:
We, Edinburgh 2012 iGEM team, have improved the functionality of this part in the following way:
We prepared a plac-RFP-SacB construct which can be used for assessing counter-selection efficiency. Part:BBa_K917002
We placed sacB under the lac promoter which allows IPTG dependent control rather than kanamycin dependent control and IPTG concentration-dependent control of the levels of selection.
We added RFP to allow distinguishing loss of the counter-selection cassette form loss of SacB fuction.
We prepared Kan-plac-RFP-sacB selection-counterselection cassette Part:BBa_K917010
Improving use of sacB for negative selection in liquid culture
The 2011 Wisconsin-Madison team utilized and tested this part as part of their double selection cassette. It was used to perform directed evolution upon different liquid phase fluorescent E. coli biosensors. In light of their findings, they have suggested improvements to the protocol for use of this part.
For the 2011 Wisconsin-Madison team's project, they required the selection to be done in liquid culture, as opposed to the solid agar plate use shown in the BRIDGE protocol by the 2010 Edinburgh team above. This allowed for testing of a large mutant library that would have been impractical to screen on plates. Additionally, they were testing E. coli biosensors designed to work with liquid analytes which were impossible to use in a solid agar plate (ethanol and alkanes).
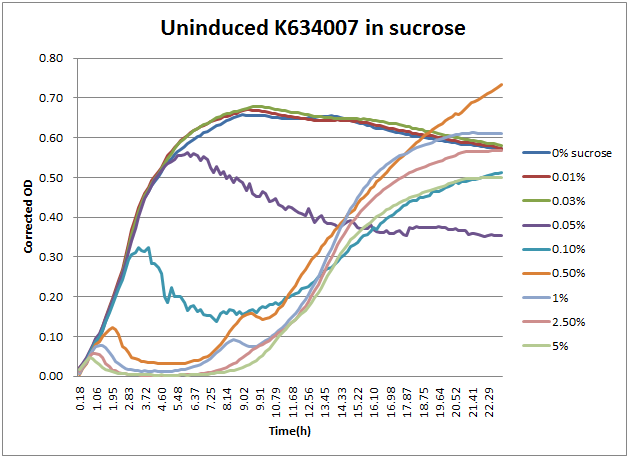
As can be seen above, the expression of sacB in the presence of sucrose prevents most growth on a solid plate, albeit with some survivors. In a liquid culture condition, this occasional sucrose-insensitivity presents a greater problem than sporadic colonies on a plate. When sacB-expressing cells were grown overnight in >1% v/v sucrose-containing media, the culture was always turbid in the morning. This was at first thought to reveal that sacB was not being expressed, but a time course plate reader study revealed that was not the case.
From this, the 2011 Wisconsin-Madison team proposes a new method for using K322921 for counter selections of mutant populations in liquid culture. When screening mutants that should have an effect on the expression of the sacB-containing operon, the following protocol should yield the largest possible number of mutants with reduced leaky expression. Based on the above data regarding growth in sucrose, it appears that a 5 hour liquid-culture growth would result in nearly complete death of sucrose-sensitive cells, without significant presence of sucrose-insensitive mutants. At this five hour point, the culture can either have the plasmid DNA extracted or be pelleted and plated to be screened.
The 2011 Wisconsin-Madison team hopes that this improved protocol for use of this part in liquid cultures will improve future teams' experience with this unique part. This holds great promise for high throughput screening of mutant libraries using BioBrick parts, and increases the viability of directed mutagenesis to improve existing devices.
User Reviews
UNIQdfec59b6c433175c-partinfo-00000000-QINU UNIQdfec59b6c433175c-partinfo-00000001-QINU