Part:BBa_K731201
Arabinose inducible araC-pBAD promoter
This E. coli promoter is positively regulated by arabinose.
araC is the regulator protein: in absence of arabinose the protein binds to araI1 and araO2 and the DNA forms a loop that prevents RNA polymerase from binding the promoter. When arabinose is added to the culture it binds to araC, disabling the interaction with araI1 and araO2 sites, and also allowing it to bind araI site, promoting transcription.
Promoter activity is also influenced by glucose levels: in its absence cAMP levels are high and the the complex cAMP+CAP (catabolite activator protein) binds to lacI promoting transcription. In presence of glucose, cAMP levels are low and promoter activity is inhibited.
This part differs in several point mutations from part BBa_I0500, mainly in araC coding sequence.
Usage and Biology
This part has been successfully operated both in pSB1C3 (K731030) and the low copy vector pSB3C5, in which it was characterized.
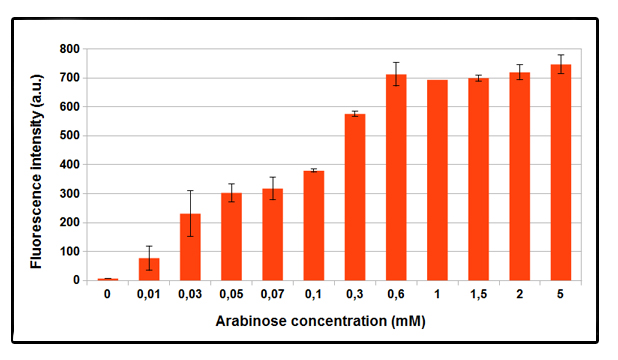
FIGURE 1. GFP expression with different concentrations of arabinose, after 4 hours of induction.
Test was conducted in NEB10beta strain, with part BBa_K731250, but in pSB3C5. Cells were grown in LB broth and induced at OD_600 = 0.6 . Higher GFP expression is observed as the arabinose concentration increases. No difference in fluorescence intensity between not induced and control NEB10beta cells was observed. Data shown is a mean of three different measurements.
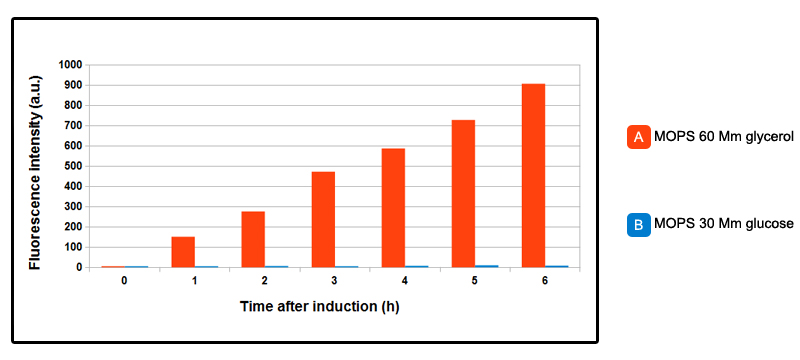
FIGURE 2. GFP expression over time, after induction.
Test was conducted in NEB10beta strain, with part BBa_K731250, but in pSB3C5. Cells were grown in LB brothuntil OD_600= 0.3 then centrifuged and resuspended in equal amount of minimal MOPS medium and induced at OD_600 = 0.6 with 5mM arabinose concentration. MOPS A is 60mM glycerol as carbon source, MOPS B is 30mM glucose: in the chart, data relative to MOPS A are shown in orange, the ones relative MOPS B are in blue. In absence of glucose, increasing linear expression over time of GFP is observed, while in MOPS B, in presence of glucose, promoter is inhibited, and fluorescence levels are comparable with control NEB10beta cells. In a later experiment done with a superfolderGFP-tagged CysE enzyme, some fluorescence in presence of glucose was observed. You can see the results on this page.
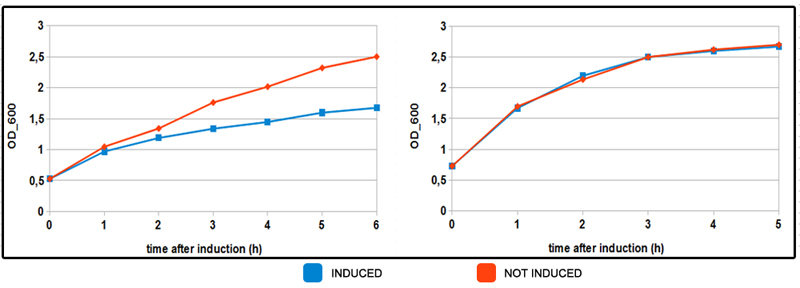
FIGURE 3. Induced and not induced culture growth in MOPS mediums.
Test was conducted in NEB10beta strain, with part BBa_K731250, but in pSB3C5. Cells were grown in LB brothuntil OD_600= 0.3 then centrifuged and resuspended in equal amount of minimal MOPS medium and induced at OD_600 = 0.6 with 5mM arabinose concentration. On the left OD mesurements of coltures growing in minimal MOPS A medium (glycerol, 60mM). In orange not induced culture, in blue induced (arabinose 5mM). Same colors are used on the chart on the right for induced-not induced bacteria with MOPS B medium (30 mM glucose). Inhibition of growth is observed in MOPS A when induced. There seem to be no effect in MOPS B. This particular behaviour is probably due to energy assessment while growing on glycerol as sole carbon source. More energy is invested in protein production as observed in FIGURE 2.
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21INCOMPATIBLE WITH RFC[21]Illegal BamHI site found at 1144
- 23COMPATIBLE WITH RFC[23]
- 25INCOMPATIBLE WITH RFC[25]Illegal AgeI site found at 979
- 1000INCOMPATIBLE WITH RFC[1000]Illegal SapI site found at 961
//direction/forward
//chassis/prokaryote/ecoli
//promoter
//regulation/positive
//classic/regulatory/other
| None |