Coding
cspB
Part:BBa_K525123
Designed by: Anna Drong Group: iGEM11_Bielefeld-Germany (2011-09-11)
Revision as of 16:22, 21 September 2011 by JSchwarzhans (Talk | contribs)
S-layer cspB from Corynebacterium glutamicum with lipid anchor and PT7 and RBS
Sequence and Features
Assembly Compatibility:
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21INCOMPATIBLE WITH RFC[21]Illegal BamHI site found at 1334
Illegal XhoI site found at 161
Illegal XhoI site found at 779 - 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000INCOMPATIBLE WITH RFC[1000]Illegal BsaI.rc site found at 1313
Illegal SapI site found at 560
Illegal SapI site found at 772
Illegal SapI site found at 1320
Expression in E. coli
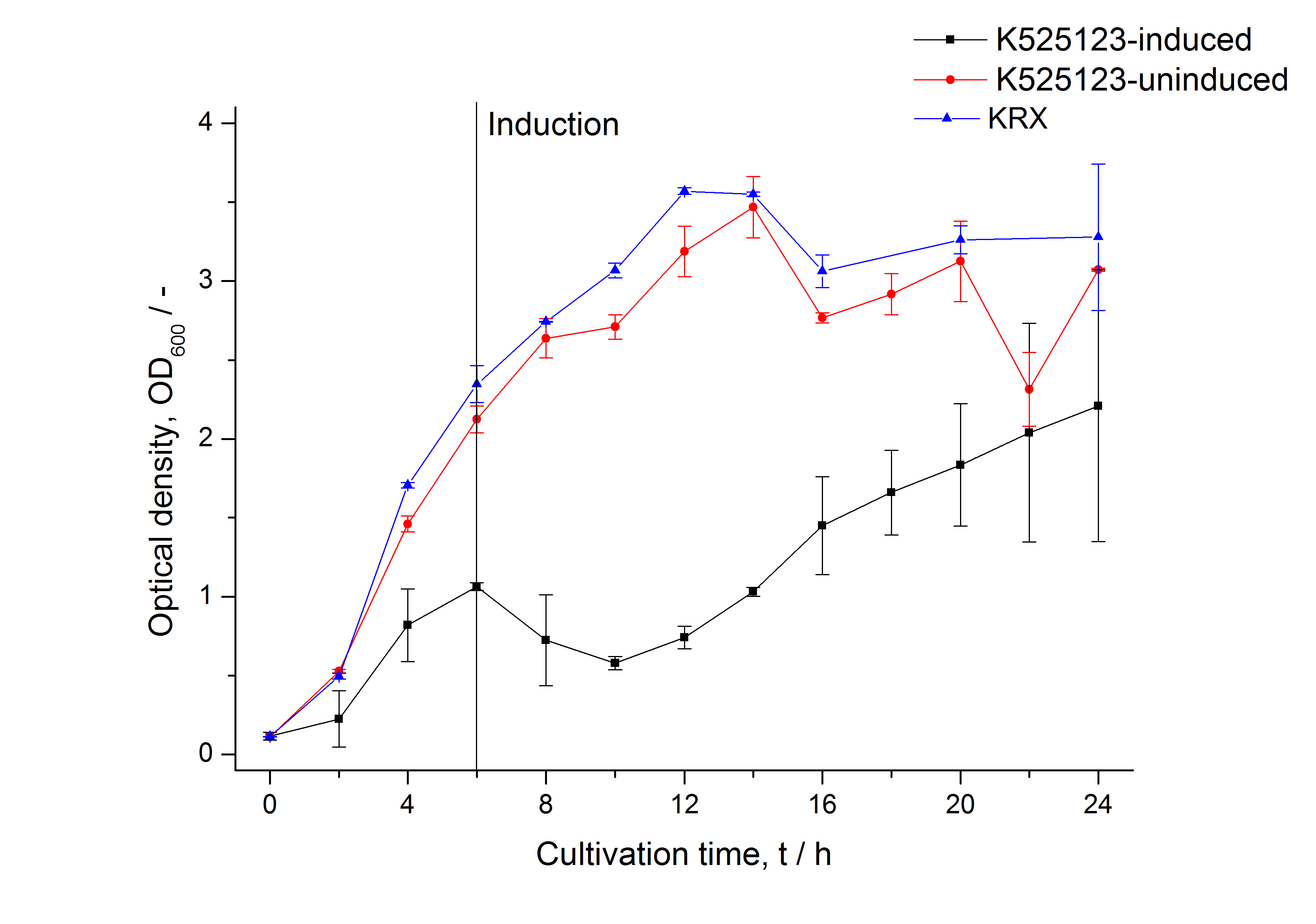
The CspB gen was fused with a monomeric RFP (BBa_E1010) using [http://2011.igem.org/Team:Bielefeld-Germany/Protocols#Gibson_assembly Gibson assembly] for characterization.
The CspB|mRFP fusion protein was overexpressed in E. coli KRX after induction of T7 polymerase by supplementation of 0,1 % L-rhamnose using the [http://2011.igem.org/Team:Bielefeld-Germany/Protocols/Downstream-processing#Expression_of_S-layer_genes_in_E._coli autinduction protocol] from promega.
[edit]
Categories
Parameters
//chassis/prokaryote/ecoli
//proteindomain/internal
//rnap/bacteriophage/t7
//proteindomain/internal
//rnap/bacteriophage/t7
| n/a | S-layer cspB from Corynebacterium glutamicum with lipid anchor and PT7 and RBS |