Part:BBa_K2010999
PETase (PET-degrading enzyme, origin I. sakaiensis)
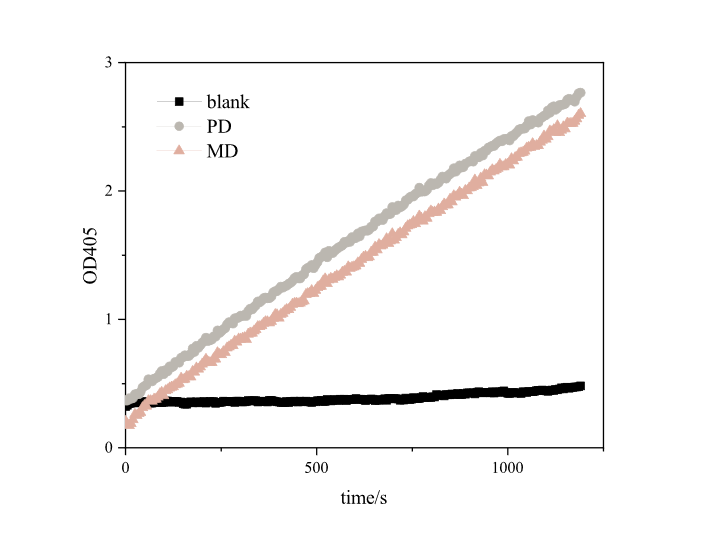
PETase is a poly(ethylene terehphthalate)-degrading enzyme first identified in Ideonella sakaiensis. BBa_K2010999 is the sequence for the E. coli K12 optimized DNA sequence for PETase. The catalytic activity of IsPETase can be characterized using a p-nitrophenol butyrate (pNPB) assay, in which PETase cleaves the ester bond of pNPB to release p-nitrophenolate. The absorbance of this compound can be measured at 405 nm to reflect IsPETase activity.

This SDS-PAGE gel demonstrates successful protein purification of PETase. PETase has a molecular weight of approximately 30 kDa.

This graph demonstrates PETase activity with various substrate concentrations at a pH of 7.0.

This graph demonstrates PETase activity with various substrate concentrations at a pH of 8.0.

This graph demonstrates PETase activity with various substrate concentrations at a pH of 9.0.

This graph demonstrates PETase activity as a function of pH. PETase is most active at a pH of 9.0, but activity at 8.0 and 9.0 is practically indistinguishable.
Contribution
Group: KEYSTONE 2020
Summary: According to a study by Brandon C. Knott et al., PETase can be linked with MHETase in an two-enzyme system to increase the efficiency of degradation significantly. The deconstruction of recalcitrant polymers is done in nature by synergistic enzyme cocktails. There is a recent discovery on a type of two-enzyme system for the structure of polyethylene terephthalate(PET). This system used a kind of enzyme to transform the polymer to a soluble intermediate and another kind of enzyme for constituent PET monomer production. This discovery suggests that nature may start to involve with the deconstruction of synthetic plastics. The evolution of microbes is with the ability to use synthetic polymers as sources of carbon and energy. Ideonella sakaiensis was found recently to secrete a two-enzyme system. The sakaiensis PETase depolymerizes PET, and release soluble products, such as 2-hydroxyethy, MHET that was cleaved from the terephthalic acid and ethylene glycol by MHETase. 1.6 Å resolution MHETase structure was recorded, which means MHETase core domain is covered by a lid domain and has similarity with PETase. Simulations of catalytic itinerary predict the MHETase uses the typical two-step serine hydrolase mechanism. The Bioinformatics analysis said the MHETase is a result of the evolution of ferulic acid esterases, and the two homologous enzymes show the exhibit of MHET turnover. The result of two homologous enzymes and MHETase S131G also shows the residue is very important for the accommodation of MHET about the active site. MHETase lid is also important to hydrolysis of MHET, and MHETase does not turnover mono-furanoate of mono-isophalate. Also, all exhibit improved PET and MHET show turnover to free enzymes of the PETase chimeric proteins of linker lengths. These results suggested information for future work on the two-enzyme PET depolymerization, and can contribute to future deconstruction of plastics through biological ways.
References: Brandon C. Knott (13 Oct. 2020.). Characterization and engineering of a two-enzyme system for plastics depolymerization. PNAS. Retrieved from https://www.pnas.org/content/117/41/25476
Contribution
Group: Austin UTexas
Summary: PETase’s ability to depolymerize PET is limited by its low stability in environmental conditions. However, PETase has increased stability when displayed through a yeast cell-surface display system designed by Chen et al. This group found that PETase is most stable at a pH of 9 and a temperature of 40 °C. Below this temperature conformation changes lead to reduced stability. Crystallinity was also found to affect PETase’s function in plastic-degradation. PETase is less effective at degrading samples of PET with more crystallized molecular structures. These results also indicated that protein concentration has a great influence on the turnover rate of PETase. Increasing protein concentration over a threshold causes enzyme oversaturation, since not all PETase molecules can react with a given sample of PET at once.1
References: Chen, Z., Wang, Y., Cheng, Y., Wang, X., Tong, S., Yang, H., & Wang, Z. (2020). Efficient biodegradation of highly crystallized polyethylene terephthalate through cell surface display of bacterial PETase. Science Of The Total Environment, 709, 136138. doi: 10.1016/j.scitotenv.2019.136138
Contribution
Group: Uppsala 2022
Summary: Poly(ethylene terephthalate) (PET) hydrolysing enzymes like PETase with the ability to depolymerise this highly abundant plastic type gained major interest after their discovery as they posess great potential of green and scalable way of recycling even in industrial scale. Major drawbacks so far have been their low activity, instability to pH and temperature ranges and inability to use untreated PET (1). To overcome those problems engineering work on the enzyme PETase originally found in ideonella sakaiensis (2) was documented within the iGEM competition as well as in scientific journals. One of the latest Nature publications working with PETase used a machine learning-aided engineering approach with the goal to improve the overall enzyme stability (3). In total they found 4 mutations S121E, T140D, R224Q and N233K which highly increased the stability both individually and in combination. They further used these findings to evaluate the hydrolytic activity of their newly generated stable mutants. In all their generated mutants they identified one mutant called FAST-PETase (functional, active, stable and tolerant PETase) which had the highest overall activity at 50°C compared to all other known PET hydrolysing enzymes. FAST-PETase which contained the mutations S121E, D186H, R224Q, N233K and R280A performed well compared to other known enzymes in temperatures between 30, 40 °C and a pH range of 6.5-8 which represent moderate conditions which should be relatively easy to sustain on larger scale. Structural analysis revealed that the increased stability of this mutant as being due to the N233K mutation which places a positively charged lysine residue next to a negatively charged glutamic acid which results in a salt bridge. The R224Q mutation allows the glutamine to form a hydrogen bond to the carbonyl group of a neighboring serine. Finally the S121E mutation allows a new established water-mediated hydrogen-bonding network with a histidine and asparagine.
References:
1. Taniguchi, I. et al. Biodegradation of PET: current status and application aspects. ACS Catal. https://doi.org/10.1021/acscatal.8b05171 (2019).
2. Yoshida, S. et al. A bacterium that degrades and assimilates poly(ethylene terephthalate). Science 351, 1196–1199 (2016).
3. Lu, H., Diaz, D.J., Czarnecki, N.J. et al. Machine learning-aided engineering of hydrolases for PET depolymerization. Nature 604, 662–667 (2022). https://doi.org/10.1038/s41586-022-04599-z
DUT_China 2021 improvement
Compared to the old part BBa_K2010999, expressing PETase, we design a new part BBa_K3898152, which is expressing RIDD-PETase. PETase in our research was optimized, and it shows better enzyme activity and considerable degradation effect.
Based on the results, we found that RIDD-PETase have esterase activity. After 20 minutes of reaction, each resulted in absorption peaks as high as 3.4873 at 405nm. Our optimized RIDD-PETase enzyme activity is 1.5 times higher than theirs in BBa_K2010999.
XHD-WuHan-Pro-China 2023
Description
The increasing commodification of modern society and the close association of plastic with various industries, according to research statistics, result in the global production of approximately 100 million tons of plastic products annually, with a year-on-year increase [1]. Especially in 2020, during the fight against the COVID-19 pandemic, products primarily made from plastic materials such as gloves, masks, and protective suits played a crucial role in controlling the spread of the virus [2]. Polyethylene terephthalate (PET), due to its excellent mechanical properties and practicality, has become one of the widely used plastic materials [3], with global PET production capacity surpassing 100 million tons in 2020 [4].
Currently, PET is applied in various fields such as beverage or mineral water bottles, films, and polyester clothing [5], resulting in a significant amount of PET waste. While many countries have started recycling PET waste, the quantity remains low, and most discarded PET is not effectively recycled. These materials undergo embrittlement due to atmospheric ultraviolet radiation, free radical oxidation, and seawater hydrolysis, resulting in microscopic plastic particles that are invisible to the naked eye. These microplastics can be absorbed by aquatic organisms, and as humans sit at the top of the food chain, the ingestion of these aquatic organisms may lead to the accumulation of a substantial amount of microplastics in the human body, posing unpredictable health risks [6].
As is well-known, microorganisms play a significant role in the removal of pollutants and material cycling in ecosystems. Since the 1990s, the application of microbial enzymes in the degradation of high-molecular-weight materials has begun to draw attention as a unique solution to address the environmental issues caused by plastics. From the discovery of the first PET-degrading enzyme in the thermophilic actinomycete Thermobifida fusca by Müller and others in 2005 [7], several research teams have conducted a series of studies on PET-degrading enzymes from various microbial sources. Recently, researchers in Japan found a bacterium (Ideonella sakaiensis) capable of degrading PET and isolated an enzyme from it called PETase. This enzyme has the ability to break down hcPET (high crystallinity PET), effectively dismantling its long-chain molecules, enabling degradation [8].
Usage and Biology
To study the expression and function of PETase in Escherichia coli Rosetta, we synthesized the PETase gene (Azenta life science, USA) and cloned it into the pET23b vector, allowing the target protein to be continuously expressed in E. coli Rosetta without the need for IPTG induction. The recombinant vector was transformed into E. coli Rosetta cells, and positive clones were selected on LB agar plates containing ampicillin. Positive clones were verified by DNA sequencing (Generalbiol, China).

Figure 1 Design of gene circuit of PETase overexpression system.
Characterization
To verify the PET enzyme activity, the engineered E. coli were cultured overnight in LB medium, and cell pellets were collected by centrifugation and resuspended in Tris-HCl (pH 7.4). Cells were then lysed by sonication (150 W, sonication for 1s, interval 3s, for a total of 20 minutes) to obtain cell lysate. The total protein concentration in the lysate was measured using a Bradford reagent kit (Beyotime, China). Enzyme activity was measured using para-nitrophenyl butyrate (pNPB) as a substrate. A 100 mM pNPB stock solution (Merck, Germany) was prepared in acetonitrile. PETase activity was tested in a 100 μL reaction system: 1 mM pNPB, 50 mM Tris-HCl buffer (pH 7.4), 20 μL crude enzyme solution. The reaction was carried out at 30°C for 30 min. The release of para-nitrophenol ester from PETase cleavage of pNPB was measured at 405 nm using a microplate reader. The content of pNPB was calculated based on a standard curve. One enzyme activity unit (U) is defined as the amount of enzyme required to release 1 μmol of para-nitrophenol ester per minute. The specific activity of PETase enzyme is defined as U/mg.

Figure 2 Activity test of PETase. A: PETase enzyme kinetics curve; B: Effect of temperature on PETase activity. C:Effect of pH on the activity of PETase.
By varying the substrate pNPB concentration, we observed an increase in PETase activity. However, at a substrate concentration of 1000 μM, the enzyme activity growth rate plateaued, suggesting that the engineered bacterial strain successfully expressed PETase. Further analysis of the enzyme kinetics curve using Graphpad Prism 8.0 revealed a maximum reaction rate (Vmax) of 0.92 U/min and a Km value of 329.5 μM, calculated using the Michaelis-Menten equation (Figure 2A). Additionally, we explored the impact of temperature and pH on the microplastic processor's performance. The results indicated that the processor effectively operated within a temperature range of 25°C to 45°C, with optimal efficiency observed at 40°C. For pH, the processor functioned well within the pH range of 7.4 to 9.8, with peak efficiency at pH 9.2 (see Figure 2B and 2C).
Potential application directions
We have confirmed the successful expression and characterization of PETase enzyme in engineered Escherichia coli. The enzyme's ability to degrade PET suggests promising potential applications in the bioremediation of microplastic pollution in various environments, particularly aquatic ecosystems. Furthermore, the enzyme operates effectively within a temperature range of 25°C to 45°C, with peak performance observed at 37°C, making it suitable for diverse environmental conditions. This includes potential utilization in wastewater treatment plants, where the enzyme can assist in breaking down PET-based contaminants.
The capability of the microplastic processor to function efficiently in a slightly alkaline environment (pH 7.4 to 9.8) implies its suitability for addressing microplastic pollution in seawater without the need for additional pH adjustments, which is particularly valuable for marine conservation efforts. The research findings may lay the groundwork for bioengineering approaches to develop specialized microorganisms or enzyme-based solutions for targeted microplastic cleanup in ecosystems significantly affected by plastic pollution.
References
[1] QUECHOLAC-PIÑA X, GARCÌA-RIVERA MA, ESPINOSA-VALDEMAR RM, VÁZQUEZ-MORILLAS A, BELTRÁN-VILLAVICENCIO M, CISNEROS�RAMOS AL. Biodegradation of compostable and oxodegradable plastic films by backyard composting and bioaugmentation[J]. Environmental Science and Pollution Research International, 2017, 24(33): 25725-25730. [2] WONG SL, NGADI N, ABDULLAH TAT, INUWA IM. Current state and future prospects of plastic waste as source of fuel: a review[J]. Renewable and Sustainable Energy Reviews, 2015, 50: 1167-1180. [3] ROCHMAN CM, BROWNE MA, HALPERN BS, HENTSCHEL BT, HOH E, KARAPANAGIOTI HK, RIOS-MENDOZA LM, TAKADA H, TEH S, THOMPSON RC. Policy: classify plastic waste as hazardous[J]. Nature, 2013, 494(7436): 169-171. [4] GEYER R, JAMBECK JR, LAW KL. Production, use, and fate of all plastics ever made[J]. Science Advances, 2017, 3(7): e1700782-e1700782. [5] MOHARIR RV, KUMAR S. Challenges associated with plastic waste disposal and allied microbial routes for its effective degradation: a comprehensive review[J]. Journal of Cleaner Production, 2019, 208: 65-76. [6] WEBB H, ARNOTT J, CRAWFORD R, IVANOVA E. Plastic degradation and its environmental implications with special reference to poly(ethylene terephthalate)[J]. Polymers, 2012, 5(1): 1-18. [7] WIERCKX N, PRIETO MA, POMPOSIELLO P, de LORENZO V, O’CONNOR K, BLANK LM. Plastic waste as a novel substrate for industrial biotechnology[J]. Microbial Biotechnology, 2015, 8(6): 900-903. [8] CHEN, ZHOUZI, et al. "Efficient biodegradation of highly crystallized polyethylene terephthalate through cell surface display of bacterial PETase." Science of The Total Environment 709 (2020): 136138.
- 10COMPATIBLE WITH RFC[10]
- 12INCOMPATIBLE WITH RFC[12]Illegal NotI site found at 67
- 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000COMPATIBLE WITH RFC[1000]
| None |