Part:BBa_K4800001
rrnB T1 terminator
Carboxylate reductase from Mycobacterium marinum(Mutation of glutamine at position 302 to glutamate)
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000COMPATIBLE WITH RFC[1000]
Conclusion : We performed Time of Flight Mass Spectrometer on the purified HIS-tagged amajLime protein. The predicted molecular mass of this protein is about 26840Da. The result of TOF-Mass Spectrometry showed that the specific molecular mass of amajLime protein is 26.898kDa (the value of the sharpest peak is shown as the molecular mass of amajLime protein). Moreover, the intensity of 26.898kDa is up to 4x105, which indicates the high concentration and purity of the amajLime protein. There are also some small protein peaks, suggesting that the noise had some effect, but not much.
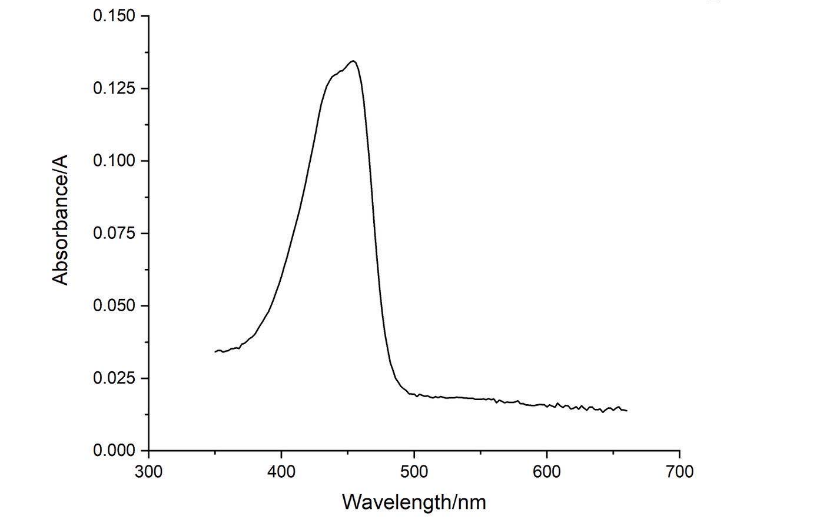
amajLime protein full-wavelength scan profile :
1-198nm 2.551A
2-210nm 2.683A
3-276nm 0.146A
4-454nm 0.135A
Conclusion : The full-wavelength scan of amajLime protein shows that the strongest absorption peak of amajLime protein occurs at 210nm. As shown in the results, amajLime has a low intensity peak at 400 to 450 nm, which may be due to the fluorescence excitation demonstrated by previous teams such as Hong Kong-CUHK iGEM 2017.
Conclusion : The full wavelength measurement of amajLime (359-660nm) was compared with the excitation spectrum figure of 2013UPPSALA , indicating that the results of amajLime protein characterized by our team were similar to those of 2013 UPPSALA and Hong Kong-CUHK iGEM 2017.
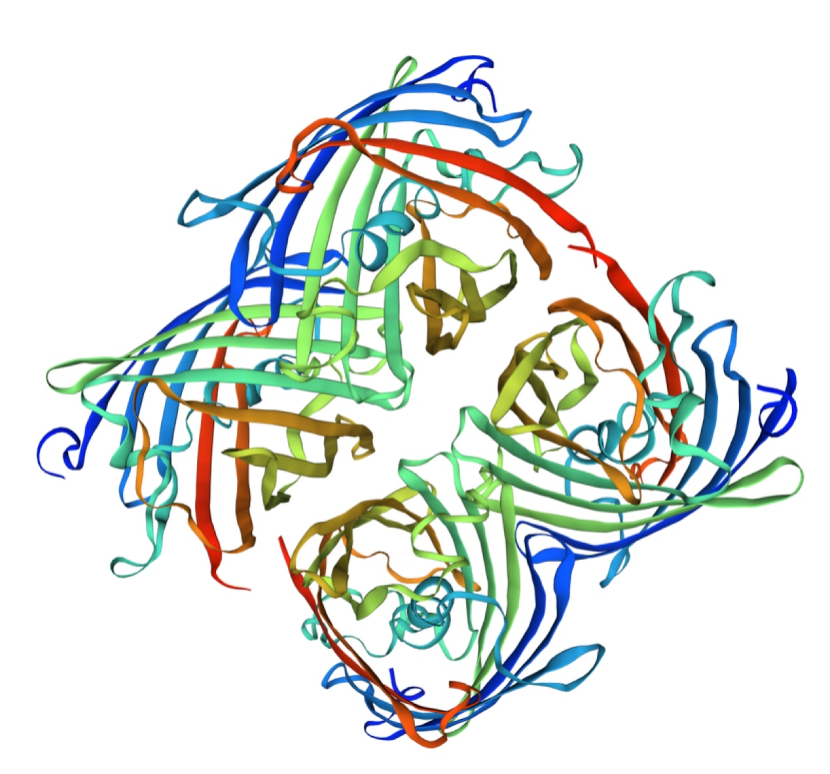
Structural modeling results of the amajLime protein based on Swiss-Model
Conclusion: We used Swiss-Model to simulate the three-dimensional structure of amajLime protein. The above figures showed the modeling result of Swiss-Model.
| None |