Part:BBa_K4275013
OlpB-3C-Ag3
OlpB3CAg3, an outer layer protein B with 3 type II cohesins and an antigen 3 (Ag3), is the secondary scaffoldin of cellulosome complex. The main function of OlpB is securing cellulosome integrating protein A (CipA) to the cell surface. The type II cohesin interacts with the type II dockerin on CipA, binding CipA to OlpB. In turn, OlpB adheres to the cell surface through the interaction between Ag3 and nanobody 3 (Nb3) present on the cell surface. The binding to the cell surface effectively increases the efficiency of the cellulosome complex. This is a part in a part collection where we enable efficient degradation of cellulose and PET in textile waste.
Figure 1 The 3D structure of the protein predicted by Alphafold2.
Usage and Biology
OlpB3CAg3 is the secondary scaffoldin of cellulosome complex that has 3 type II cohesins and an antigen 3 (Ag3). Natural OlpB is found in Clostridium thermocellum, a gram-positive thermophilic and anaerobic bacterium. OlpB is responsible for binding several primary scaffoldin of cellulosome complex, CipA, to the cell surface [1]. OlpB binds to CipA via the interaction between the type II dockerin and type II cohesin. In nature, OlpB attaches to the cell surface of Clostridium thermocellum through the interaction between surface-like homologous (SLH) domains and the cell surface. Together, the CipA1B2C, OlpB, and the cellulosomal enzymes comprise a multiplex cellulosome. Utilizing the highly effective cellulosome complex, C.thermocellum is the most efficient microorganism for lignocellulosic biomass degradation.
Characterisation of the Cellulosome Complex
Mini-scaffold construction of cellulosome
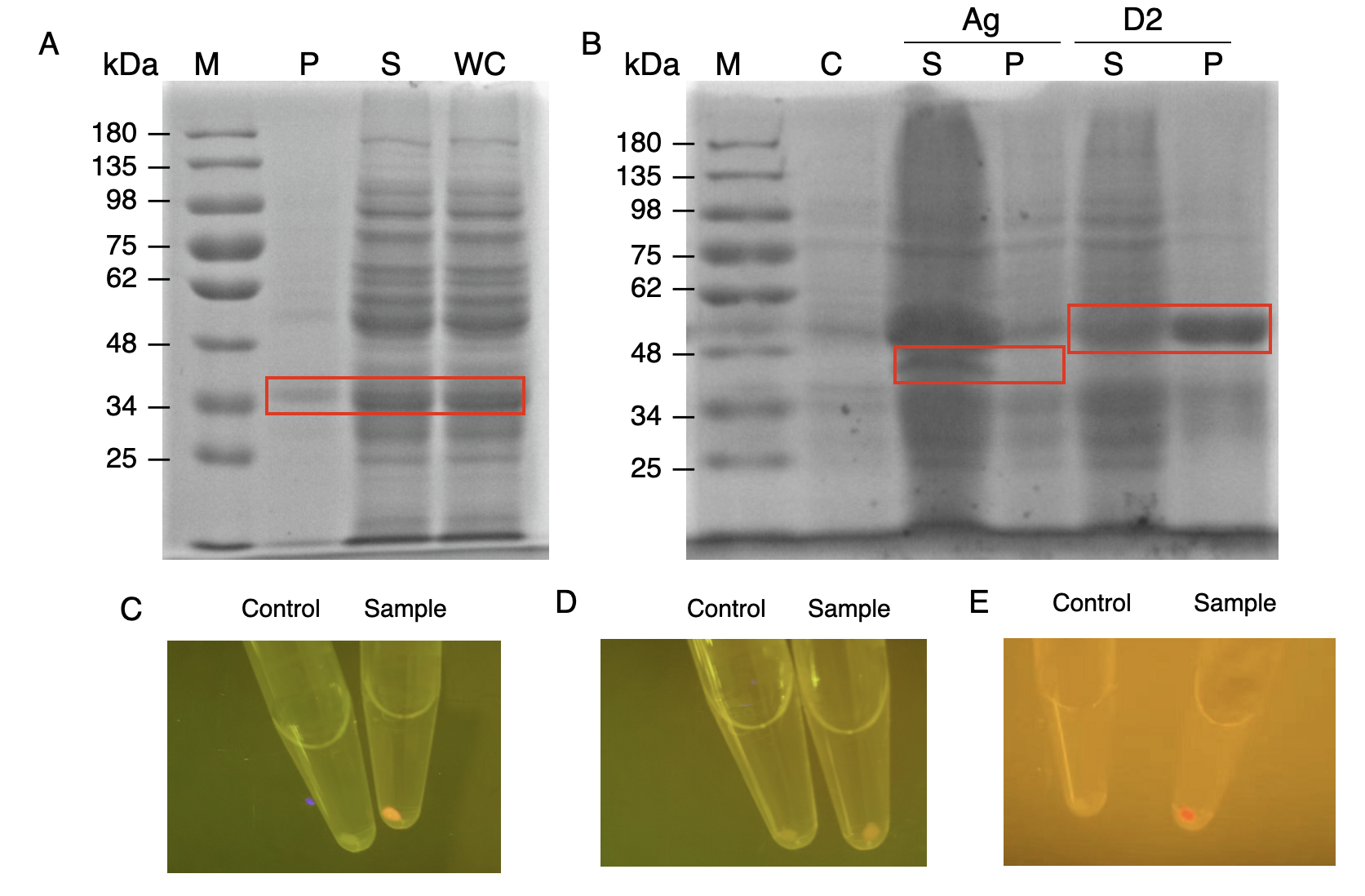
We constructed E.coli expression vectors for the mini-scaffold protein subunits. The scaffoldin components of the wild-type cellulosome subunits are large protein scaffolds that would bring a massive protein burden to the bacterial host secreting them. We modified the coding sequences for the wild-type cellulosome protein scaffold, as shown in (Fig.2A and Fig.2B) The mini-scaffolds were successfully expressed by our host, verified by the SDS-PAGE analysis shown in (Fig.2C and Fig.2D)
Functionality testing of our mini-scaffold
In order to verify the three levels of protein-protein interaction that assembles our cellulosome complex, Ag3-eforRED, DocI-eforRED, and DocII-eforRED vectors were constructed and cultured for IPTG-inducible expression (Fig. 3D). SDS-page analysis was performed with lysed cells and all three targeted proteins were identified in both whole cell and supernatant (Fig. 4A and 4B).
The nanobody-antigen interaction was verified by mixing intact E. coli cells displaying Neae-Nb3 with the supernatant of Ag3-eforRED (Fig. 3A). Red fluorescent characteristics were observed in the pellets after resuspending the centrifuged mixture, which is absent in the control group that only contains Neae-Nb3 (Fig. 4C).
After that, the type II cohesin-dockerin interaction was tested using the mixture of Neae-Nb3, OlpB-Ag3, and the type II dokerin fused with eforRED (Fig. 3B). A negative control lacking OlpB-Ag3 was set up for result comparison. Centrifugation was used to remove supernatant and the red fluorescence was only identified in pellets of the sample group, confirming the type II cohesin-dockerin interaction (Fig. 4D).
Finally, the association between type I cohesin and type I dockerin was validated using the mixture of Neae-Nb3, OlpB-Ag3, CipA1B2C, and DocI-eforRED (Fig. 3C), red fluorescence was detected in the resuspended mixture while it was not observed in the control group lacking the primary scaffold CipA1B2C (Fig. 4E), verifying the type I cohesin-dockerin interaction.
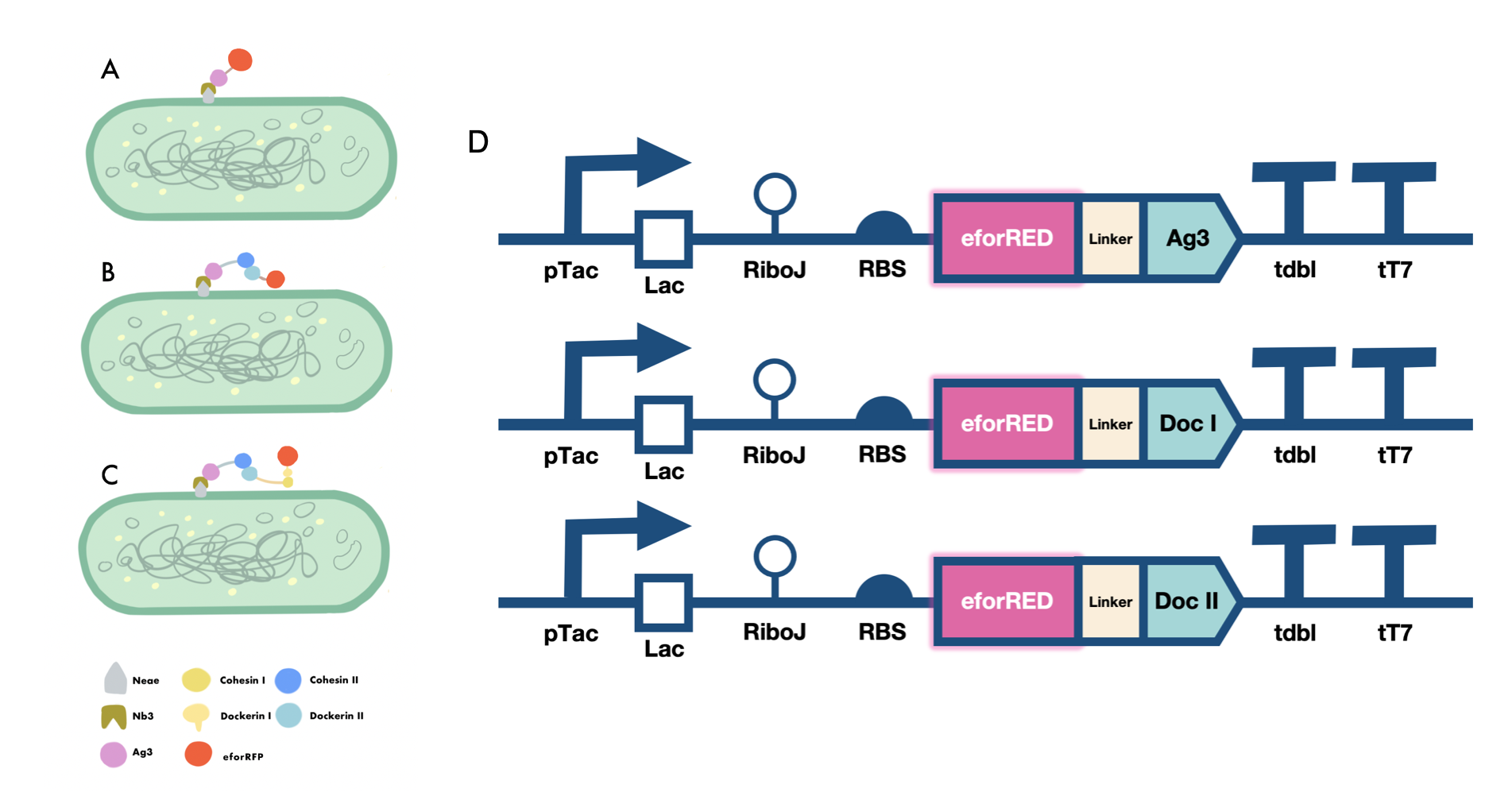
Cellulosome construction
We assembled the cellulose-like complex on the surface of E.coli by adding primary scaffold proteins, cellulases and cellulase boosters onto E.coli expressing secondary scaffold proteins. The mixture was centrifuged and resuspended in tris-HCl. The mixture underwent centrifugation and resuspension using tris-HCl, and cellulose was added to the mixture.
After 24h, the mixture was filtered and tested for glucose by Benedict's test. From the result, we determined that the cellulosome-like complexes are able to degrade cellulose at a higher efficiency than cell-free cellulases mixture. The overall success in engineering our project was verified by the successful construction of cellulosome complex and degrading cellulose to reducing sugars.

Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21INCOMPATIBLE WITH RFC[21]Illegal XhoI site found at 706
- 23COMPATIBLE WITH RFC[23]
- 25INCOMPATIBLE WITH RFC[25]Illegal NgoMIV site found at 1252
Illegal NgoMIV site found at 1341
Illegal AgeI site found at 183
Illegal AgeI site found at 319
Illegal AgeI site found at 370
Illegal AgeI site found at 502
Illegal AgeI site found at 868
Illegal AgeI site found at 886
Illegal AgeI site found at 1312 - 1000INCOMPATIBLE WITH RFC[1000]Illegal SapI.rc site found at 1069
Illegal SapI.rc site found at 1081
Illegal SapI.rc site found at 1705
References
1. Anandharaj, Marimuthu et al. "Constructing A Yeast To Express The Largest Cellulosome Complex On The Cell Surface". Proceedings Of The National Academy Of Sciences, vol 117, no. 5, 2020, pp. 2385-2394. Proceedings Of The National Academy Of Sciences, https://doi.org/10.1073/pnas.1916529117.
| None |


