Part:BBa_K4439013
mSA-GFP-CBD-10xHis
Contents
Abstract
To complete
Sequence and features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000COMPATIBLE WITH RFC[1000]
Protein Characterization
Usage and Biology
Modeling
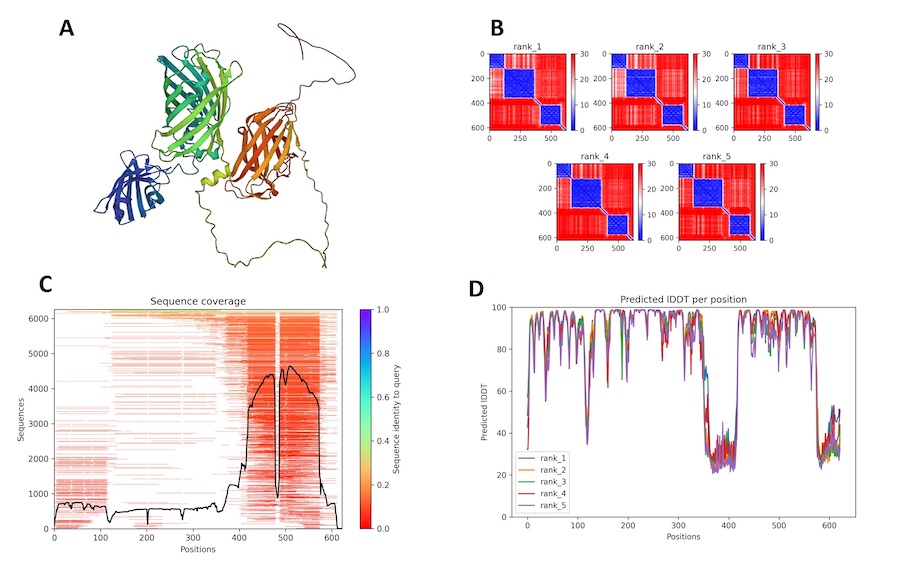
Figure 9 : AlphaFold2 prediction for 03a. (A) First rank 3D model prediction of 03a protein; the iteration who got the highest score in the modeling. (B) Different correlation graphs between the query sequence of 03a and the predicted one, for each proposed 5 models. (C) Figure of sequence coverage of 03a and indices on alignment with other sequences in the mSA. (D) IDDT graph for 03a per residue to get an idea of the confidence in the model in predicting.
- Analysis
In (fig.9, A), we could identify the three main parts of the mSA-GFP-CBD protein. Compared to the previous (fig.9, A) we could see that the GFP domain, in green, is more easily characterized and would yield a closer linkage to the mSA chain, in blue. So we will have both of the proteins closely associated. However for the CBD region, we identified a longer linkage which would mean different configuration of tha attachment possible. From (fig.9, B, D) it was really interesting to see that the 5 predictions yielded similar correlation measures and similar IDDT scores, which enabled us to confirm that the model is robust in the determination of three chain structures. The linkage would give in reality more freedom of placement of those chains.
Results
Bacterial Transformation
Protein Purification
Cloning by PCR and KLD
References
| None |
