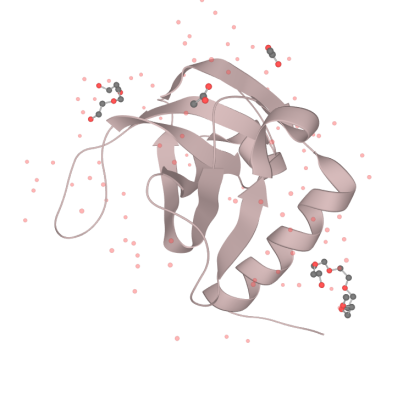
Part:BBa_K3331005
nodA(MazF)
ndoA (ydcD, mazF) [ Bacillus subtilis subsp. subtilis str. 168 ]
Encodes EndoA, an endoribonuclease that inactivates cellular mRNAs by cleaving them at specific, but frequently occurring sites. EndoA has similar cleavage specificity to MazF and PemK and yields cleavage products with 3′‐phosphate and 5′‐hydroxyl groups, typical of EDTA‐resistant degradative RNases.
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000COMPATIBLE WITH RFC[1000]
Characterization
This year, our team designed a suicide switch mediated by arabinose operon for biosafety. We use MazF to kill the bacteria. We have supplemented the relevant information of this element in the iGEM Parts and added test and characterization to it.
Our MazF comes from the genome of Bacillus Subtilis. By regulating the expression of MazF, we inhibit the life of bacteria when needed to prevent the possible strains and genetic leakage. MazF is a kind of mRNA interferas. Specifically, it specifically cleaves five-base U^ACAU sequence (^indicating the cleavage site) on RNA[1]. MazF achieves its catalytic function by forming a special MazF-RNA complex. One molecule of 9-mer RNA is bound to one MazF dimer, and two subunits of MazF form the dimer related by local 2-fold symmetry.The RNA in an extended alignment is bound along the RNA binding interface between subunits of the MazF dimer, covering part of this dimeric interface, and MazF interacts extensively with the pentad target sequence present in the RNA chain by interactions with bases[2]. Therefore, the backbone phosphate moieties project outward and away from the protein surface. In this way, scissile phosphate will be attacked by other side chain groups, which causes breaking of P-O bond.
What’s more, it is an important part of the Bacillus Subtilis toxin-antitoxin system. Which is essential for the programmed cell death of this developmental bacterium. In normally growing cells, MazF forms a stable complex with its cognate antitoxin, MazE, however, under stress conditions, unstable MazE is preferentially degraded to release free MazF in the cells, which then cleaves cellular mRNAs to inhibit protein synthesis, leading to growth arrest. The protein-protein interaction interface between MazE and MazF (2,843 Å2) is larger than the MazF-RNA interaction interface (2,153 Å2), suggesting that MazF is likely to have a higher affinity for MazE over its RNA substrate. In the B.S MazE-MazF complex structure, MazE binds to MazF along the dimer interface, and it even occupies part of the putative modeled RNA binding site on the second subunit of MazF[3]. Thus, in the presence of MazE, MazF cannot bind to or cleave substrate RNA.
Biochemical properties of MazF
divalent cation[4].
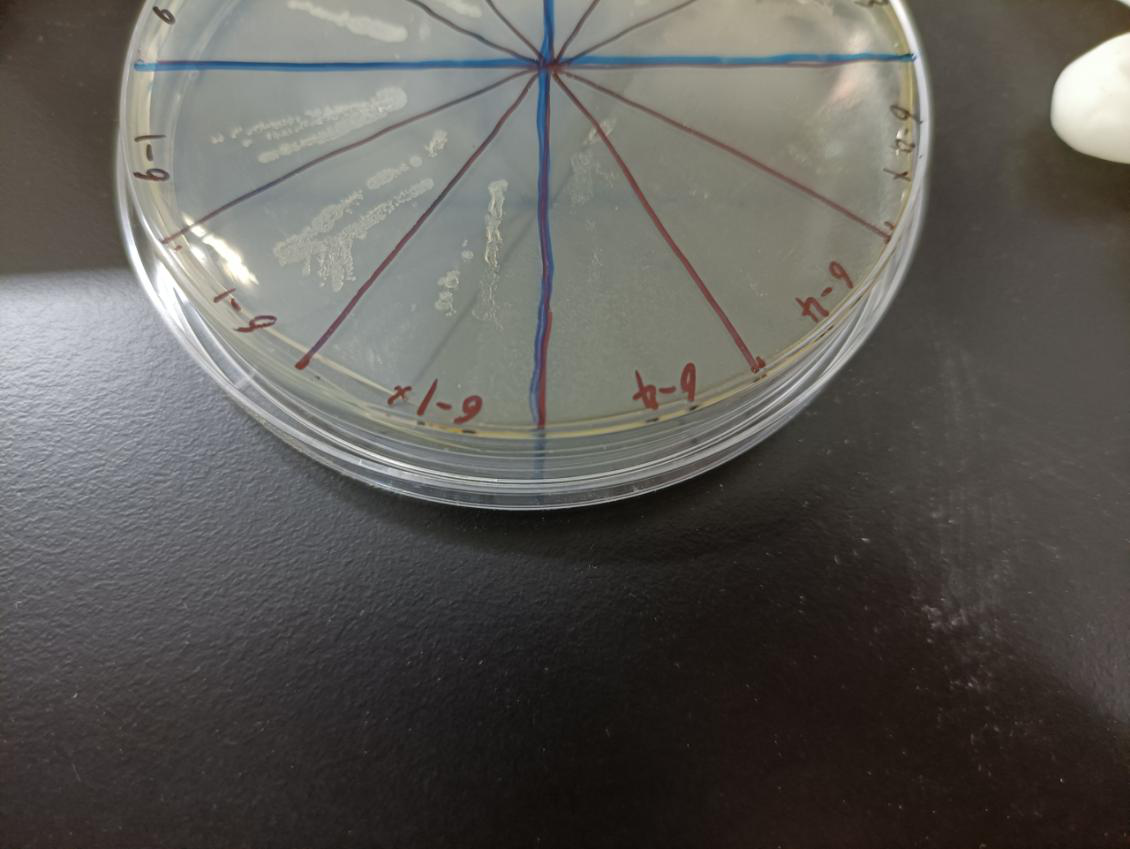
In the figure, the bacteria in this plate contained plasmids of pBAD-MazF. Three of 6-1 groups were the control group without arabinose induction, and three of 6-4 groups were the experimental group with 1.5 M/L arabinose induction. Where, the dilution factor of the group with * is 10^4, and the dilution factor of the group without * is 10^6.
Result: The bacteria without arabinose induction grow normally, while with arabinose induction, the bacteria suicide.
| None |